LINE-1 Methylation Status in Canine Splenic Hemangiosarcoma Tissue and Cell-Free DNA
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Tissue Sample DNA Extraction
2.3. Blood Collection and Cell-Free DNA Extraction
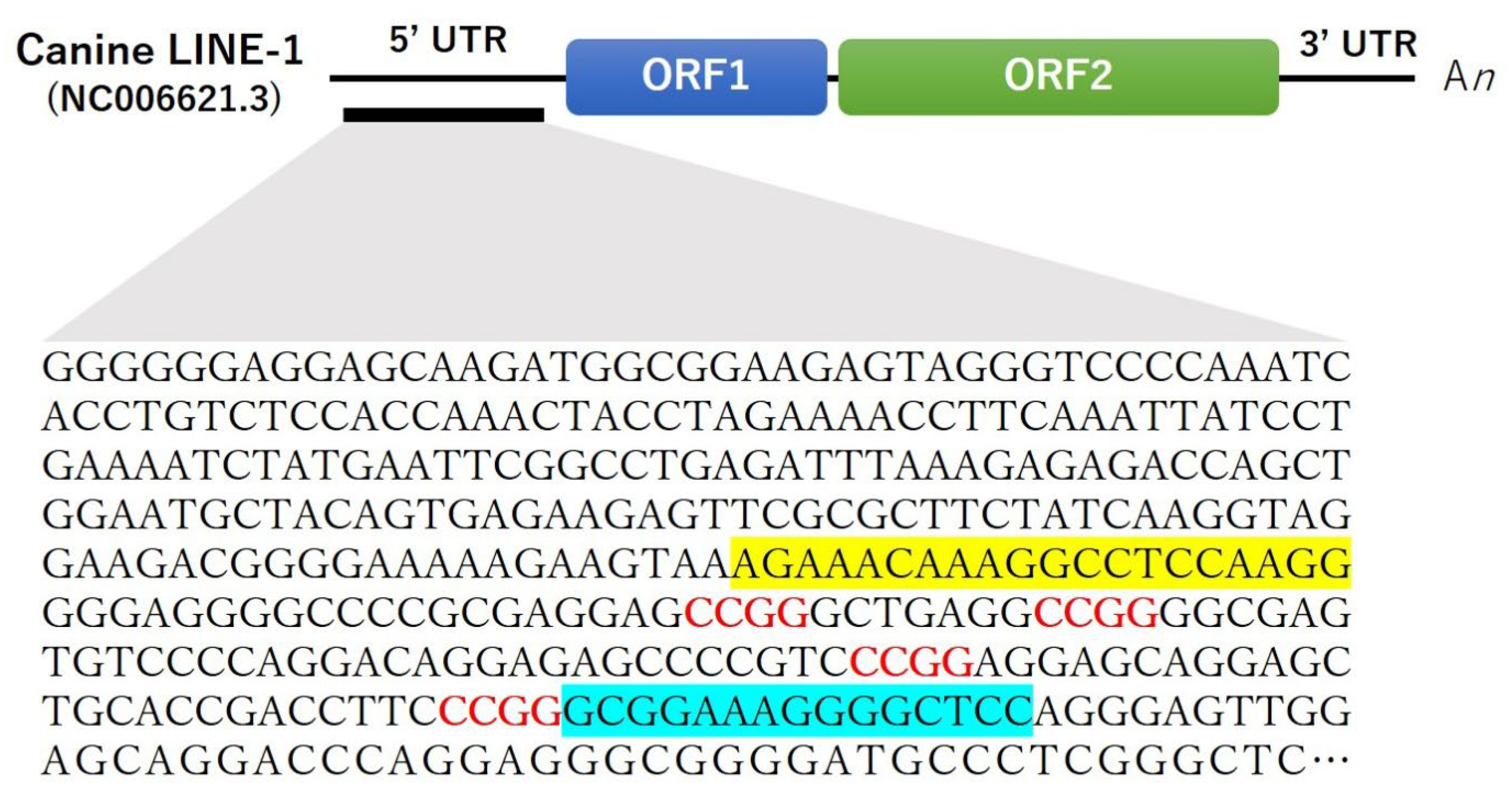
2.4. Quantification of LINE-1 Methylation
2.5. Statistical Analyses
3. Results
3.1. Background Characteristics of the Patients
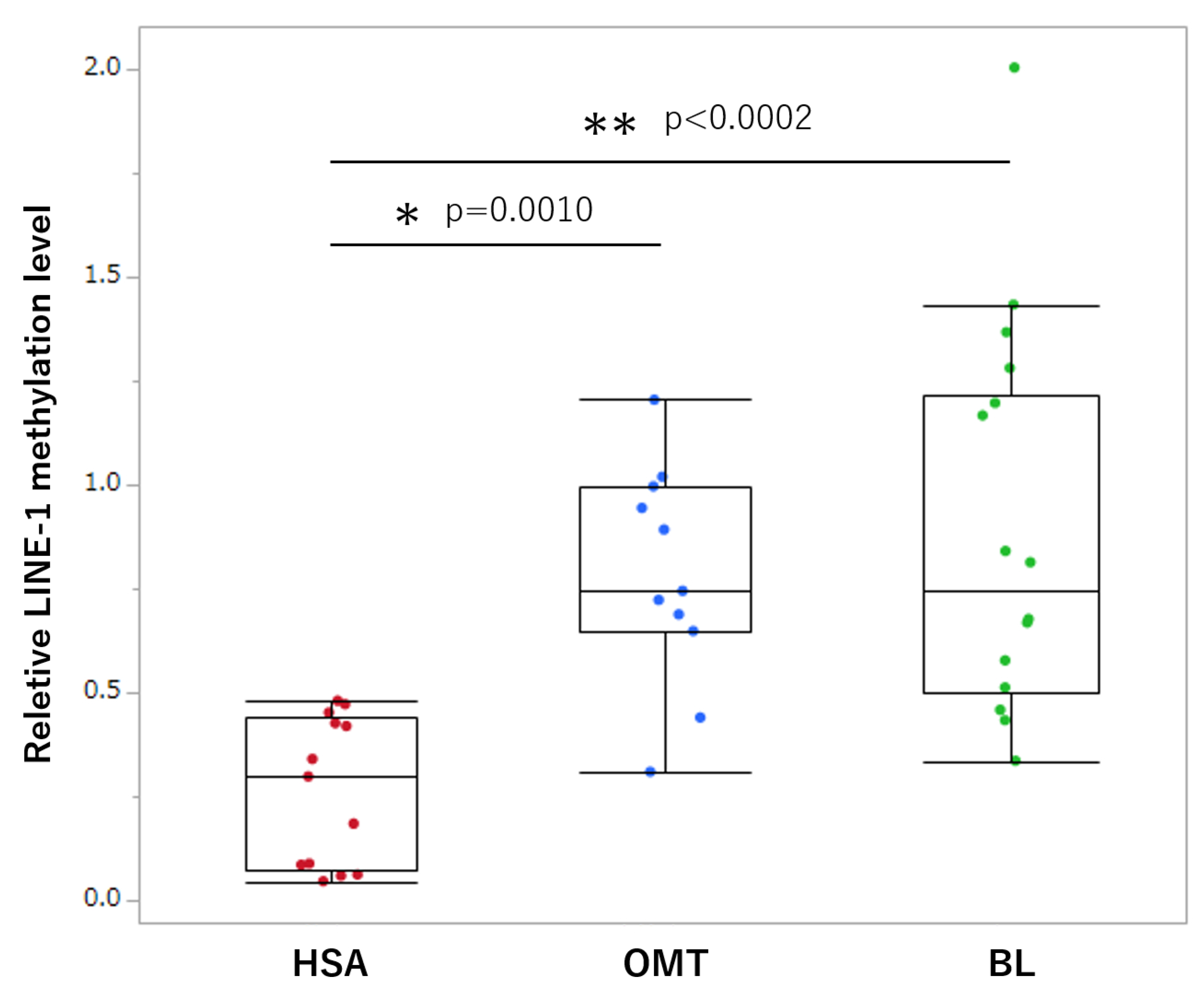
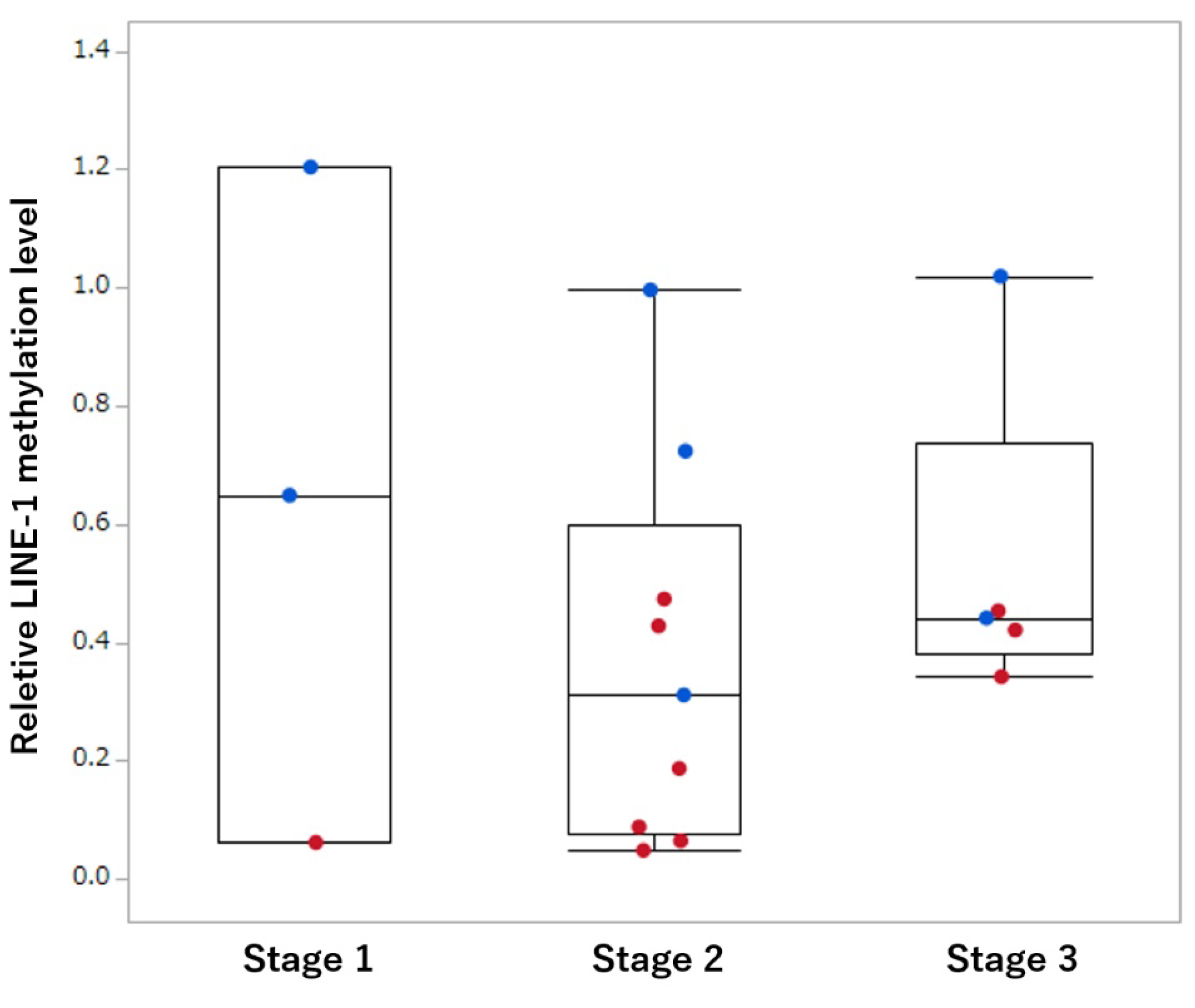
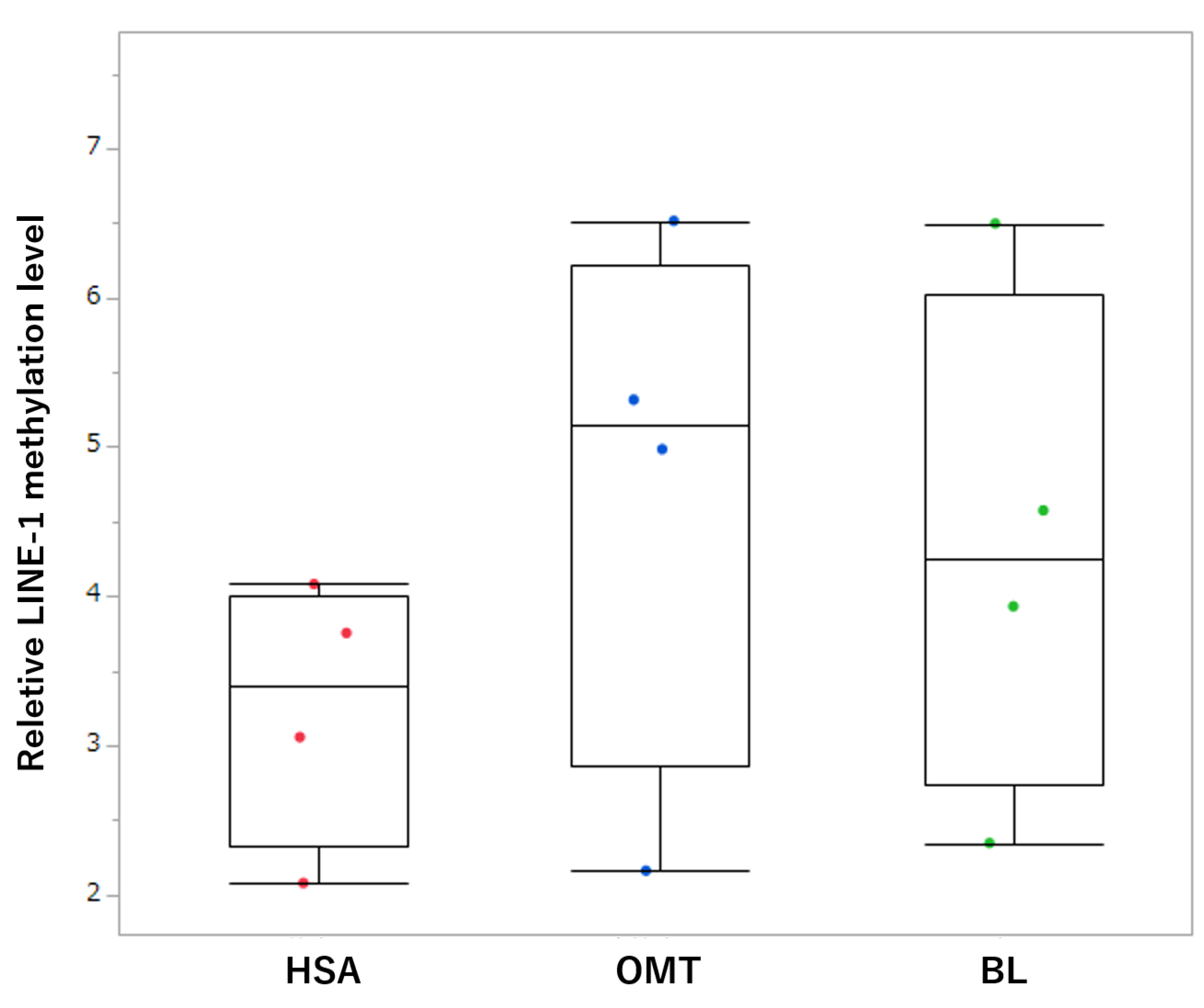
3.2. Comparison of LINE-1 Methylation in Tissue Samples
3.3. Comparison of LINE-1 Methylation in cfDNA Samples
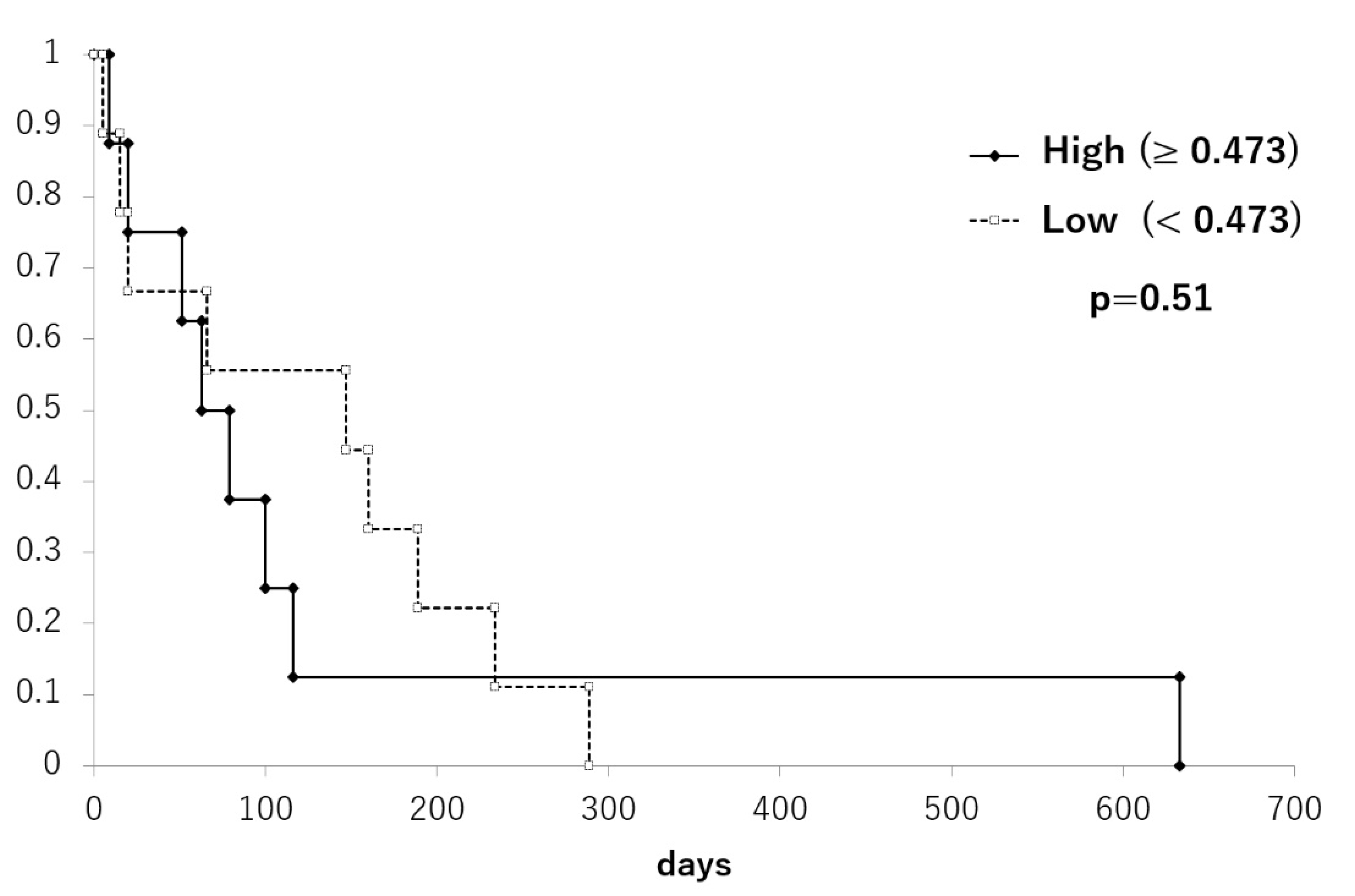
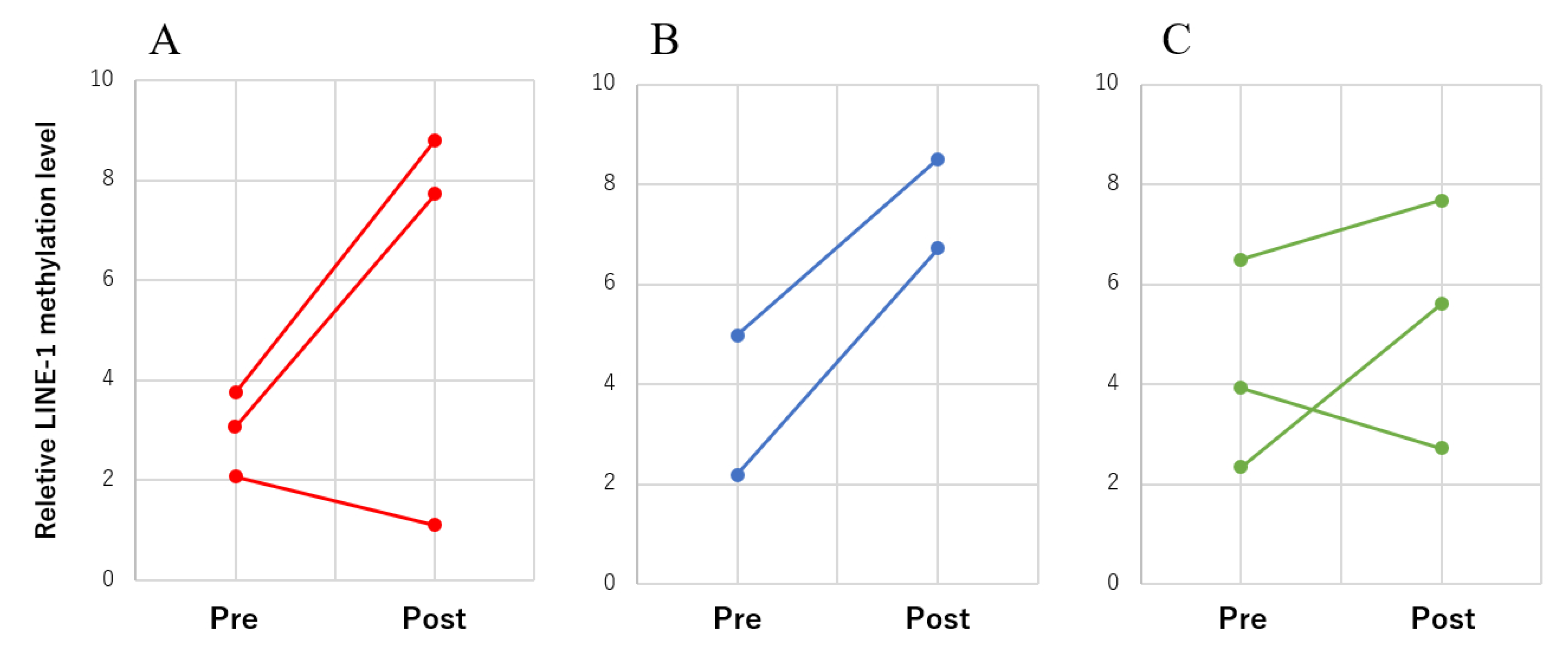
3.4. LINE-1 Methylation Changes in cfDNA during Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, K.A.; Powers, B.E.; Withrow, S.J.; Sheetz, M.J.; Curtis, C.R.; Wrigley, R.H. Splenomegaly in dogs. Predictors of neoplasia and survival after splenectomy. J. Vet. Intern. Med. 1989, 3, 160–166. [Google Scholar] [CrossRef]
- Griffin, M.A.; Culp, W.T.N.; Rebhun, R.B. Canine and feline haemangiosarcoma. Vet. Rec. 2021, 189, e585. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.O.; Patnaik, A.K.; MacEwen, E.G. Canine hemangiosarcoma: Retrospective analysis of 104 cases. J. Am. Vet. Med. Assoc. 1985, 186, 56–58. [Google Scholar]
- Wendelburg, K.M.; Price, L.L.; Burgess, K.E.; Lyons, J.A.; Lew, F.H.; Berg, J. Survival time of dogs with splenic hemangiosarcoma treated by splenectomy with or without adjuvant chemotherapy: 208 cases (2001–2012). J. Am. Vet. Med. Assoc. 2015, 247, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Spangler, W.L.; Culbertson, M.R. Prevalence, type, and importance of splenic diseases in dogs: 1480 cases (1985–1989). J. Am. Vet. Med. Assoc. 1992, 200, 829–834. [Google Scholar] [PubMed]
- Ballegeer, E.A.; Forrest, L.J.; Dickinson, R.M.; Schutten, M.M.; Delaney, F.A.; Young, K.M. Correlation of ultrasonographic appearance of lesions and cytologic and histologic diagnoses in splenic aspirates from dogs and cats: 32 cases (2002–2005). J. Am. Vet. Med. Assoc. 2007, 230, 690–696. [Google Scholar] [CrossRef]
- Lee, M.; Park, J.; Choi, H.; Lee, H.; Jeong, S.M. Presurgical assessment of splenic tumors in dogs: A retrospective study of 57 cases (2012–2017). J. Vet. Sci. 2018, 19, 827–834. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Yu, J. New Understanding of the Relevant Role of LINE-1 Retrotransposition in Human Disease and Immune Modulation. Front. Cell Dev. Biol. 2020, 8, 657. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef]
- Li, J.; Huang, Q.; Zeng, F.; Li, W.; He, Z.; Chen, W.; Zhu, Z.W.; Zhang, B. The prognostic value of global DNA hypomethylation in cancer: A meta-analysis. PLoS ONE 2014, 9, e106290. [Google Scholar] [CrossRef]
- Akimoto, N.; Zhao, M.; Ugai, T.; Zhong, R.; Lau, M.C.; Fujiyoshi, K.; Kishikawa, J.; Haruki, K.; Arima, K.; Twombly, T.S.; et al. Tumor long interspersed nucleotide element-1 (LINE-1) hypomethylation in relation to age of colorectal cancer diagnosis and prognosis. Cancers 2021, 13, 2016. [Google Scholar] [CrossRef]
- Lee, K.H.; Shin, T.J.; Kim, W.H.; Cho, J.Y. Methylation of LINE-1 in cell-free DNA serves as a liquid biopsy biomarker for human breast cancers and dog mammary tumors. Sci. Rep. 2019, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Wedge, E.; Hansen, J.W.; Garde, C.; Asmar, F.; Tholstrup, D.; Kristensen, S.S.; Munch-Petersen, H.D.; Ralfkiaer, E.; Brown, P.; Grønbaek, K.; et al. Global hypomethylation is an independent prognostic factor in diffuse large B cell lymphoma. Am. J. Hematol. 2017, 92, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Mettler, E.; Fottner, C.; Bakhshandeh, N.; Trenkler, A.; Kuchen, R.; Weber, M.M. Quantitative analysis of plasma cell-free DNA and its DNA integrity and hypomethylation status as biomarkers for tumor burden and disease progression in patients with metastatic neuroendocrine neoplasias. Cancers 2022, 14, 1025. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, T.; Yamazaki, J.; Meagawa, S.; Yokoyama, N.; Aoshima, K.; Takiguchi, M.; Kimura, T. Long interspersed nucleotide element-1 hypomethylation in canine malignant mucosal melanoma. Vet. Comp. Oncol. 2020, 18, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Das, P.M.; Singal, R. DNA methylation and cancer. J. Clin. Oncol. 2004, 22, 4632–4642. [Google Scholar] [CrossRef]
- Li, P.; Liu, S.; Du, L.; Mohseni, G.; Zhang, Y.; Wang, C. Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin. Epigenetics 2022, 14, 118. [Google Scholar] [CrossRef]
- Siejka-Zielińska, P.; Cheng, J.; Jackson, F.; Liu, Y.; Soonawalla, Z.; Reddy, S.; Silva, M.; Puta, L.; McCain, M.V.; Culver, E.L.; et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 2021, 7, eabh0534. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, D.; Yin, Y.; Yang, T.; You, Z.; Li, D.; Chen, Y.; Jiang, Y.; Xu, S.; Geng, J.; et al. Circulating cell-free DNA-based methylation patterns for breast cancer diagnosis. NPJ Breast Cancer 2021, 7, 106. [Google Scholar] [CrossRef]
- Ting, A.H.; McGarvey, K.M.; Baylin, S.B. The cancer epigenome—Components and functional correlates. Genes Dev. 2006, 20, 3215–3231. [Google Scholar] [CrossRef]
- Saito, K.; Kawakami, K.; Matsumoto, I.; Oda, M.; Watanabe, G.; Minamoto, T. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin. Cancer Res. 2010, 16, 2418–2426. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, H.; Okamoto, T.; Shimamatsu, S.; Kohno, M.; Morodomi, Y.; Tagawa, T.; Kitao, H.; Okano, S.; Oda, Y.; Maehara, Y.; et al. LINE-1 hypomethylation is associated with malignant traits and cell proliferation in lung adenocarcinoma. Anticancer Res. 2020, 40, 5659–5666. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.L.; Hasemeier, B.; Schipper, E.; Vogel, A.; Kreipe, H.; Lehmann, U. LINE-1 hypomethylation in human hepatocellular carcinomas correlates with shorter overall survival and CIMP phenotype. PLoS ONE 2019, 14, e0216374. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, Y. Hypomethylation of the retrotransposon LINE-1 in malignancy. Jpn. J. Clin. Oncol. 2000, 30, 289–290. [Google Scholar] [CrossRef]
- Baba, Y.; Murata, A.; Watanabe, M.; Baba, H. Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer. Surg. Today 2014, 44, 1807–1816. [Google Scholar] [CrossRef]
- Kaneko, M.; Kotake, M.; Bando, H.; Yamada, T.; Takemura, H.; Minamoto, T. Prognostic and predictive significance of long interspersed nucleotide element-1 methylation in advanced-stage colorectal cancer. BMC Cancer 2016, 16, 945. [Google Scholar] [CrossRef]
- Miyata, T.; Yamashita, Y.I.; Baba, Y.; Harada, K.; Yamao, T.; Umezaki, N.; Tsukamoto, M.; Kitano, Y.; Yamamura, K.; Arima, K.; et al. Prognostic value of LINE-1 methylation level in 321 patients with primary liver cancer including hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Oncotarget 2018, 9, 20795–20806. [Google Scholar] [CrossRef][Green Version]
- Casarotto, M.; Lupato, V.; Giurato, G.; Guerrieri, R.; Sulfaro, S.; Salvati, A.; D’Angelo, E.; Furlan, C.; Menegaldo, A.; Baboci, L.; et al. LINE-1 hypomethylation is associated with poor outcomes in locoregionally advanced oropharyngeal cancer. Clin. Epigenetics 2022, 14, 171. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014, 63, 635–646. [Google Scholar] [CrossRef]
- Wong, K.; Ludwig, L.; Krijgsman, O.; Adams, D.J.; Wood, G.A.; van der Weyden, L. Comparison of the oncogenomic landscape of canine and feline hemangiosarcoma shows novel parallels with human angiosarcoma. Dis. Model Mech. 2021, 14, dmm049044. [Google Scholar] [CrossRef]
- Lin, W.H.; Xiao, J.; Ye, Z.Y.; Wei, D.L.; Zhai, X.H.; Xu, R.H.; Zeng, Z.L.; Luo, H.Y. Circulating tumor DNA methylation marker MYO1-G for diagnosis and monitoring of colorectal cancer. Clin. Epigenetics 2021, 13, 232. [Google Scholar] [CrossRef]
- Luo, H.; Wei, W.; Ye, Z.; Zheng, J.; Xu, R.H. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends. Mol. Med. 2021, 27, 482–500. [Google Scholar] [CrossRef]
- Hua, D.; Hu, Y.; Wu, Y.Y.; Cheng, Z.H.; Yu, J.; Du, X.; Huang, Z.H. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp. Mol. Pathol. 2011, 91, 455–460. [Google Scholar] [CrossRef]
- Boldrin, E.; Curtarello, M.; Dallan, M.; Alfieri, R.; Realdon, S.; Fassan, M.; Saggioro, D. Detection of LINE-1 hypomethylation in cfDNA of esophageal adenocarcinoma patients. Int. J. Mol. Sci. 2020, 21, 1547. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, N.; Mokhtari, R.; Charville, G.W.; Bui, N.; Ganjoo, K. Cutaneous angiosarcoma of the head and neck-A retrospective analysis of 47 patients. Cancers 2022, 14, 3841. [Google Scholar] [CrossRef] [PubMed]
- Weidema, M.E.; van de Geer, E.; Koelsche, C.; Desar, I.M.E.; Kemmeren, P.; Hillebrandt-Roeffen, M.H.S.; Ho, V.K.Y.; van der Graaf, W.T.A.; Versleijen-Jonkers, Y.M.H.; von Deimling, A.; et al. DNA methylation profiling identifies distinct clusters in angiosarcomas. Clin. Cancer Res. 2020, 26, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Gotea, V.; Margolin, G.; Elnitski, L. Significant associations between driver gene mutations and DNA methylation alterations across many cancer types. PLoS Comput. Biol. 2017, 13, e1005840. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | HSA (n = 13) | OMT (n = 11) | BL (n = 15) |
|---|---|---|---|
| Diagnosis (n) | HSA (13) |
|
|
| Age (years; median [range]) | 11 (8–15) | 11 (7–14) | 10 (7–13) |
| Sex | |||
| Female | 6 | 6 | 8 |
| Male | 6 | 5 | 7 |
| Unknown | 1 | - | - |
| Tumor stage a | |||
| 1 | 1 | 2 | - |
| 2 | 6 | 3 | - |
| 3 | 3 | 2 | - |
| Unknown | 3 | 4 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, H.; Watanabe, K.-I.; Kobayashi, Y.; Tomihari, M.; Uemura, A.; Tagawa, M. LINE-1 Methylation Status in Canine Splenic Hemangiosarcoma Tissue and Cell-Free DNA. Animals 2023, 13, 2987. https://doi.org/10.3390/ani13182987
Sato H, Watanabe K-I, Kobayashi Y, Tomihari M, Uemura A, Tagawa M. LINE-1 Methylation Status in Canine Splenic Hemangiosarcoma Tissue and Cell-Free DNA. Animals. 2023; 13(18):2987. https://doi.org/10.3390/ani13182987
Chicago/Turabian StyleSato, Hiroki, Ken-Ichi Watanabe, Yoshiyasu Kobayashi, Mizuki Tomihari, Akiko Uemura, and Michihito Tagawa. 2023. "LINE-1 Methylation Status in Canine Splenic Hemangiosarcoma Tissue and Cell-Free DNA" Animals 13, no. 18: 2987. https://doi.org/10.3390/ani13182987
APA StyleSato, H., Watanabe, K.-I., Kobayashi, Y., Tomihari, M., Uemura, A., & Tagawa, M. (2023). LINE-1 Methylation Status in Canine Splenic Hemangiosarcoma Tissue and Cell-Free DNA. Animals, 13(18), 2987. https://doi.org/10.3390/ani13182987





