An Exploratory Data Analysis from Ovine and Bovine RNA-Seq Identifies Pathways and Key Genes Related to Cervical Dilatation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics for Obtaining and Processing Data
2.2. Data Analysis, Identification of Gene Signatures, and Differences in Expression
3. Results
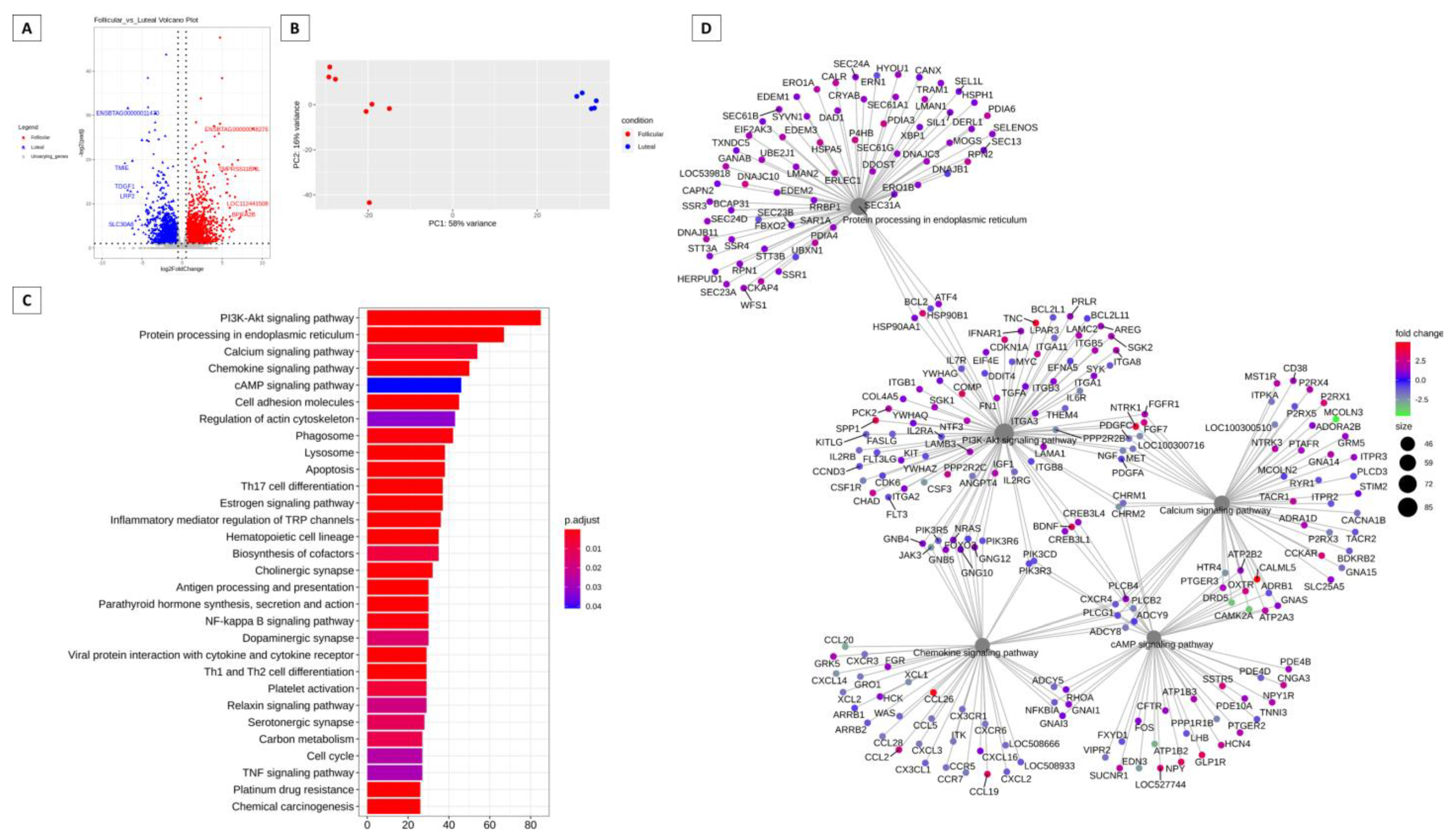
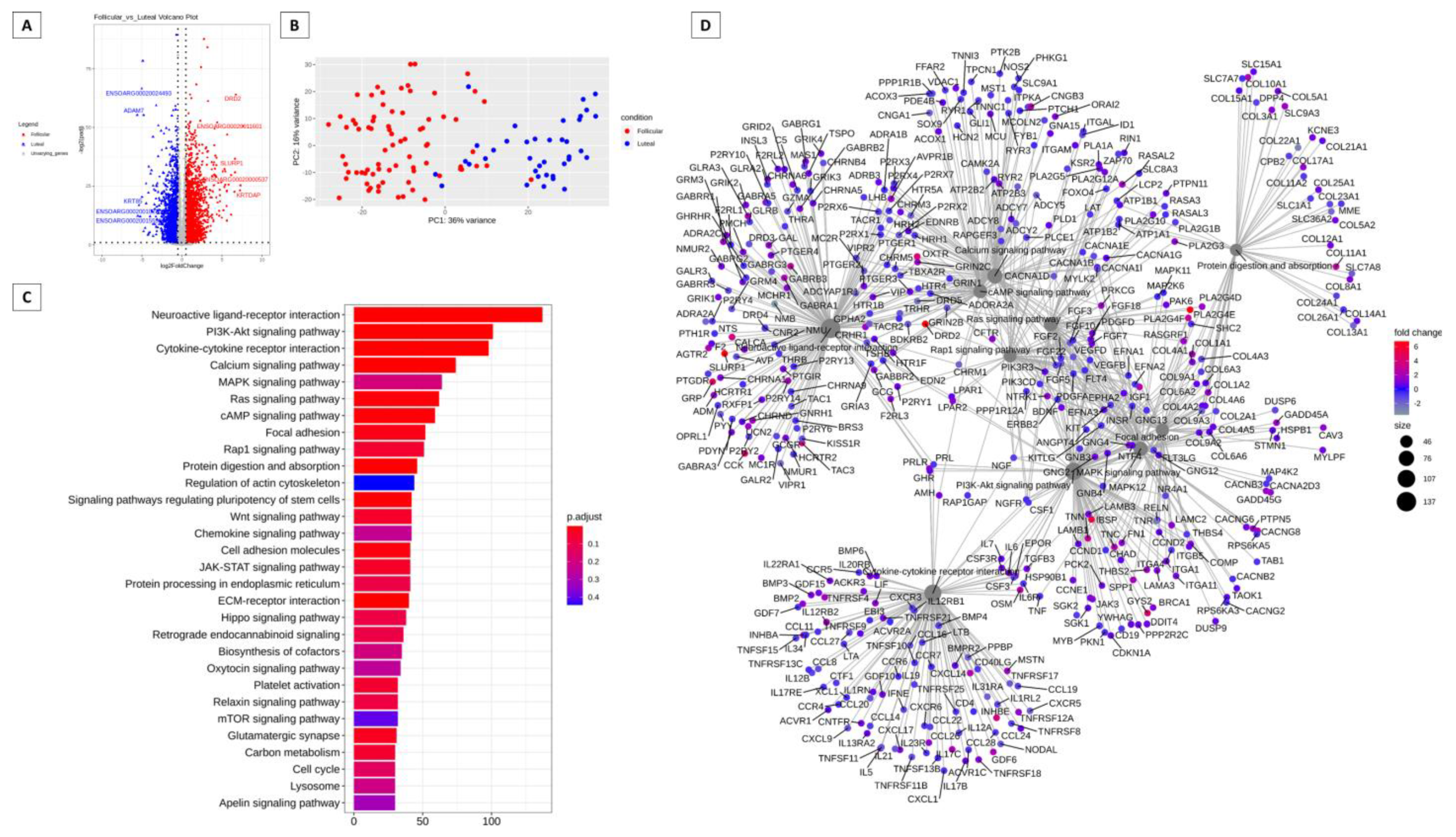
3.1. Genes More Expressed in the Follicular and Luteal Phases of Cattle and Sheep
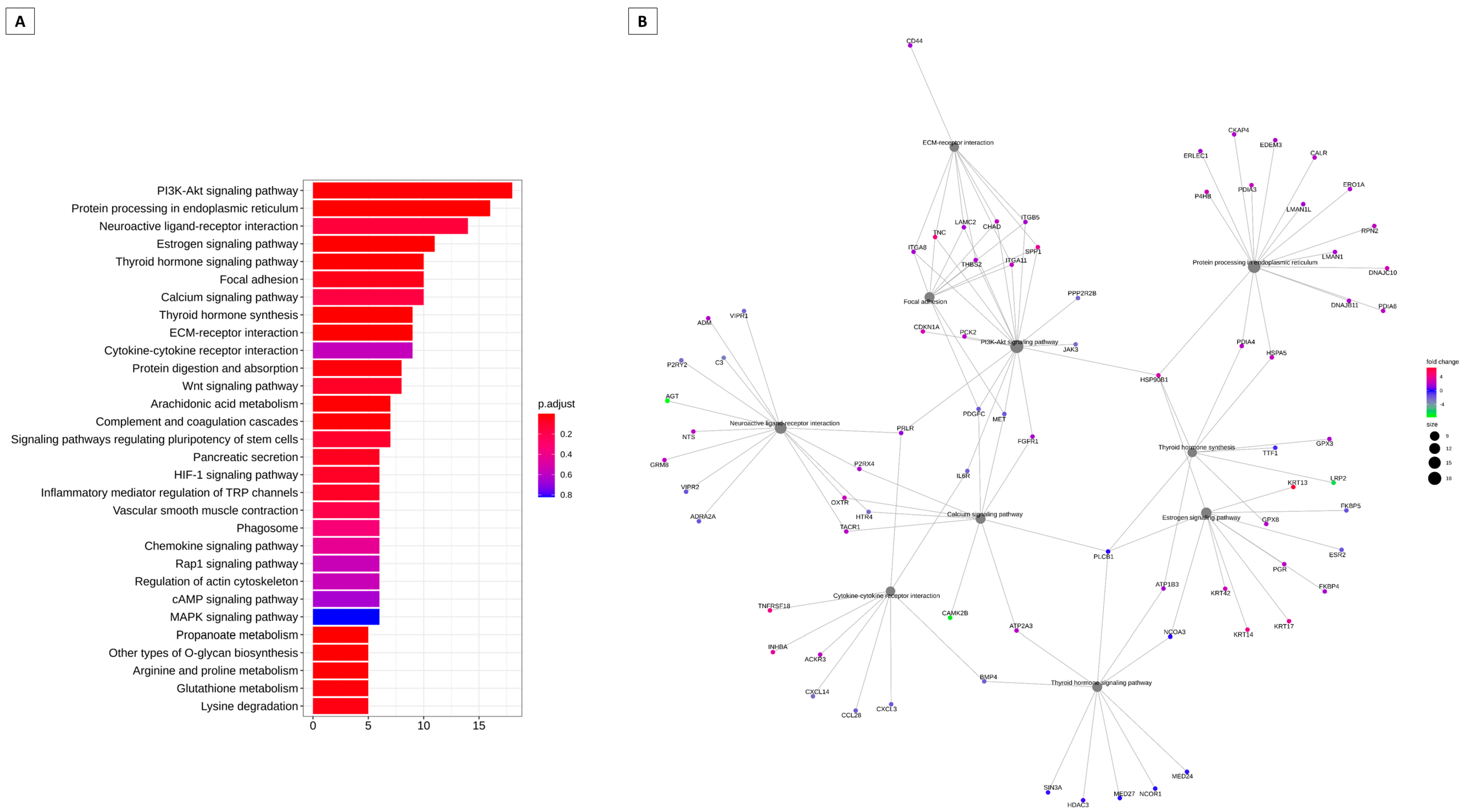
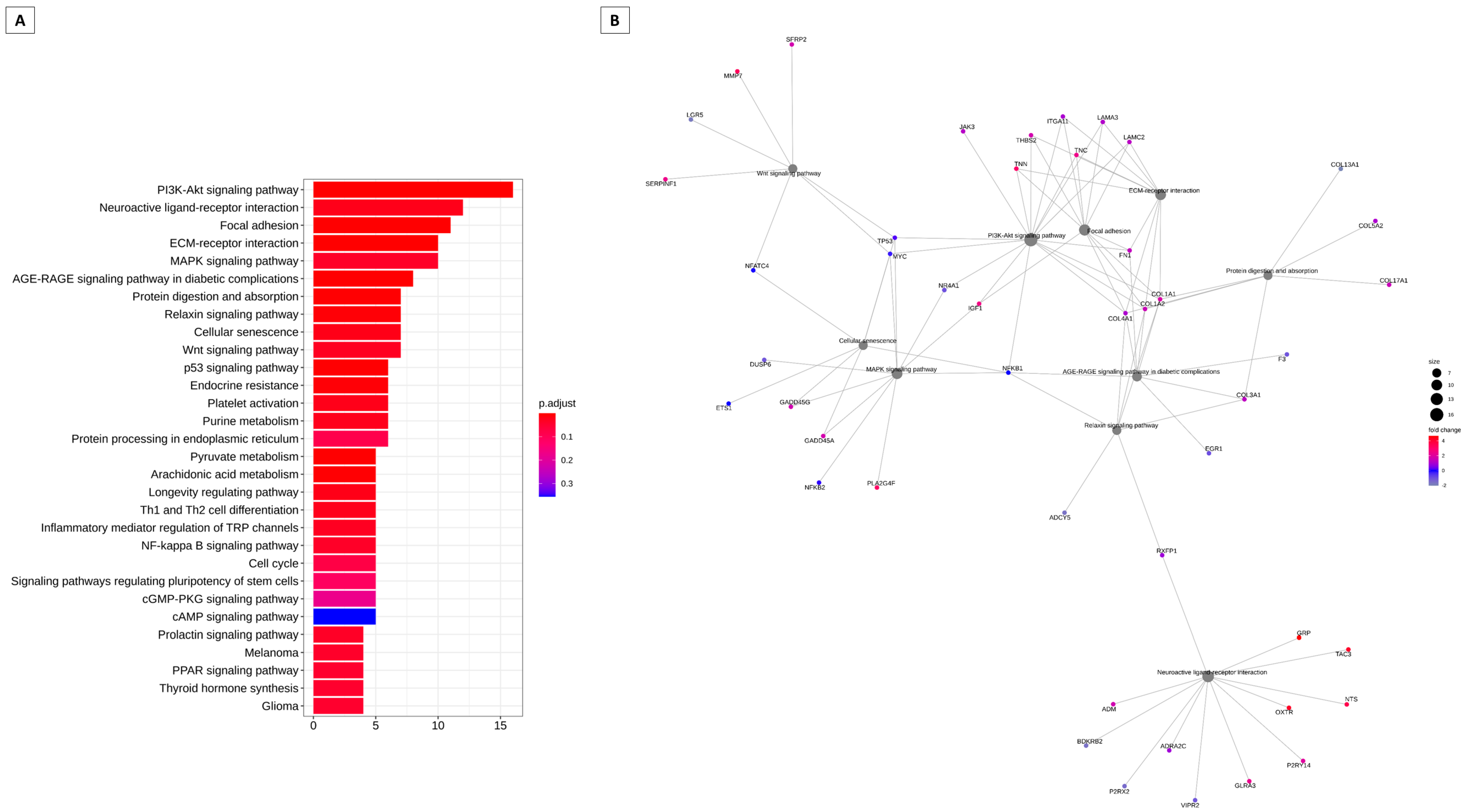
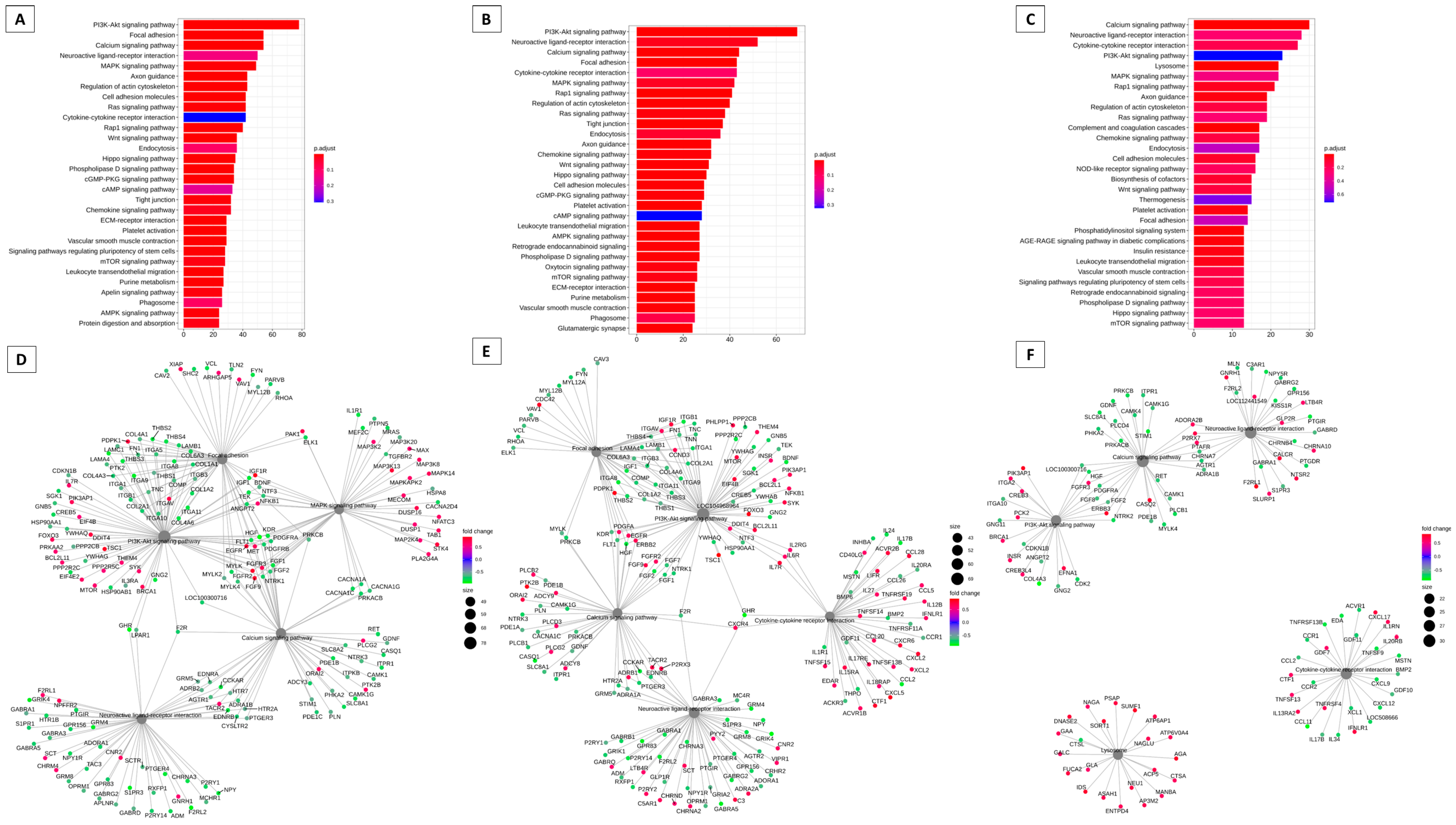
3.2. Pathways That Can Be Modulated by Key Genes
3.3. Pathways and Genes Can Be Modeled by Hox Genes
3.4. Hub Genes, 100 Most Expressed Genes and Genes Unique to the Follicular and Luteal Phase of Cattle and Sheep
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kershaw, C.M.; Khalid, M.; Mcgowan, M.R.; Ingram, K.; Leethongdee, S.; Wax, G.; Scaramuzzi, R.J. The Anatomy of the Sheep Cervix and Its Influence on the Transcervical Passage of an Inseminating Pipette into the Uterine Lumen. Theriogenology 2005, 64, 1225–1235. [Google Scholar] [CrossRef]
- Candapp, I.B.R.; Bartlewski, P.M. Induction of Cervical Dilation for Transcervical Embryo Transfer in Ewes. Reprod. Biol. Endocrinol. 2014, 12, 8. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, J.F.; Souza-Fabjan, J.M.G.; Oliveira, M.E.F.; Leite, C.R.; Nascimento-Penido, P.M.P.; Brandão, F.Z.; Lehloenya, K.C. Nonsurgical Embryo Recovery and Transfer in Sheep and Goats. Theriogenology 2016, 86, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, J.F.; Zambrini, F.N.; Guimarães, J.D.; Silva, M.R.; Oliveira, M.E.F.; Brandão, F.Z.; Bartlwwski, P.M.; Souza-Fabjan, J.M.G. Combined Treatment with Oestradiol Benzoate, D-cloprostenol and Oxytocin Permits Cervical Dilation and Nonsurgical Embryo Recovery in Ewes. Reprod. Domest. Anim. 2019, 54, 118–125. [Google Scholar] [CrossRef] [Green Version]
- Landim-Alvarenga, F.C. Parto Normal. In Obstetrícia Veterinária; Guanabara Koogan LTDA: Rio De Janeiro, Brazil, 2021. [Google Scholar]
- Nnamani, M.C.; Plaza, S.; Romero, R.; Wagner, G.P. Evidence for Independent Evolution of Functional Progesterone Withdrawal in Primates and Guinea Pigs. Evol. Med. Public Health 2013, 2013, 273–288. [Google Scholar] [CrossRef] [Green Version]
- Wagner, G.P.; Nnamani, M.C.; Chavan, A.R.; Maziarz, J.; Protopapas, S.; Condon, J.; Romero, R. Evolution of Gene Expression in the Uterine Cervix Related to Steroid Signaling: Conserved Features in the Regulation of Cervical Ripening. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mahendroo, M. Cervical Remodeling in Term and Preterm Birth: Insights from an Animal Model. Reproduction 2012, 143, 429–438. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-H.; McLenachan, P.A.; Gore, A.R.; Phillips, M.J.; Ota, R.; Hendy, M.D.; Penny, D. Four New Mitochondrial Genomes and the Increased Stability of Evolutionary Trees of Mammals from Improved Taxon Sampling. Mol. Biol. Evol. 2002, 19, 2060–2070. [Google Scholar]
- Pluta, K.; McGettigan, P.A.; Reid, C.J.; Browne, J.A.; Irwin, J.A.; Tharmalingam, T.; Corfield, A.; Baird, A.; Loftus, B.J.; Evans, A.C.O.; et al. Molecular Aspects of Mucin Biosynthesis and Mucus Formation in the Bovine Cervix during the Periestrous Period. Physiol. Genom. 2012, 44, 1165–1178. [Google Scholar]
- Abril-Parreño, L.; Meade, K.G.; Krogenæs, A.K.; Druart, X.; Fair, S.; Cormican, P. Conserved and Breed-Specific Differences in the Cervical Transcriptome of Sheep with Divergent Fertility at the Follicular Phase of a Natural Oestrus Cycle. BMC Genom. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Kassambra, A. Fastqcr: Quality Control of Sequencing Data. R package version 0.1.2.2019.
- Liao, Y.; Smyth, G.K. The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- RC Team. R: A Language and Environment for Statistical Computing; RC Team: Vienna, Austria, 2021. [Google Scholar]
- Oliveira de Biagi, C.A.; Nociti, R.P.; Brotto, D.B.; Funicheli, B.O.; Cássia Ruy, P.D.; Bianchi Ximenez, J.P.; Alves Figueiredo, D.L.; Araújo Silva, W. CeTF: An R/Bioconductor Package for Transcription Factor Co-Expression Networks Using Regulatory Impact Factors (RIF) and Partial Correlation and Information (PCIT) Analysis. BMC Genom. 2021, 22, 624. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, T.Y.; Yang, Z.Y.; Fang, W.; Wu, Q.; Zhang, C. Identification of Hub Genes in Papillary Thyroid Carcinoma: Robust Rank Aggregation and Weighted Gene Co-Expression Network Analysis. J. Transl. Med. 2020, 18, 170. [Google Scholar] [CrossRef] [Green Version]
- Lappin, T.R.J.; Grier, D.G.; Thompson, A.; Halliday, H.L. HOX GENES: Seductive Science, Mysterious Mechanisms. Ulster Med. J. 2006, 75, 23–31. [Google Scholar]
- Evans, G.; Maxwell, W.M.C. Inseminación Artificial de Ovejas y Cabras; Editorial Acribia, S.A.: España, Zaragoza, 1990; 192p. [Google Scholar]
- Masiakowski, P.; Breathnach, R.; Bloch, J.; Gannon, F.; Krust, A.; Chambon, P. Cloning of CDNA Sequences of Hormone-Regulated Genes from the MCF-7 Human Breast Cancer Cell Line. Nucleic Acids Res. 1982, 10, 7895–7903. [Google Scholar] [CrossRef] [Green Version]
- May, F.E.B.; Westley, B.R. Identification and Characterization of Estrogen-Regulated RNAs in Human Breast Cancer Cells. J. Biol. Chem. 1988, 263, 12901–12908. [Google Scholar] [CrossRef]
- Wright, N.A.; Hoffmann, W.; Otto, W.R.; Rio, M.C.; Thim, L. Rolling in the Clover: Trefoil Factor Family (TFF)-Domain Peptides, Cell Migration and Cancer. FEBS Lett. 1997, 408, 121–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taupin, D.P.D. Trefoil Factors: Initiators of Mucosal Healing. Nat. Rev. Mol. Cell Biol. 2003, 4, 721–732. [Google Scholar] [PubMed]
- Wiede, A.; Hinz, M.; Canzler, E.; Franke, K.; Quednow, C.; Hoffmann, W. Synthesis and Localization of the Mucin-Associated TFF-Peptides in the Human Uterus. Cell Tissue Res. 2001, 303, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Ruchaud-Sparagano, M.-H.; Westley, B.R.; May, F.E.B. The Trefoil Protein TFF1 Is Bound to MUC5AC in Humangastric Mucosa. Cell. Mol. Life Sci. 2004, 61, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Tomasetto, C.; Masson, R.; Linares, J.L.; Wendling, C.; Lefebvre, O.; Chenard, M.P.; Rio, M.C. PS2/TFF1 Interacts Directly with the VWFC Cysteine-Rich Domains of Mucins. Gastroenterology 2000, 118, 70–80. [Google Scholar] [CrossRef]
- Albert, T.K.; Laubinger, W.; Müller, S.; Hanisch, F.-G.; Kalinski, T.; Meyer, F.; Hoffmann, W. Human Intestinal TFF3 Forms Disulfide-Linked Heteromers with the Mucus-Associated FCGBP Protein and Is Released by Hydrogen Sulfide. J. Proteome Res. 2010, 9, 3108–3117. [Google Scholar] [CrossRef]
- Fair, S.; Hanrahan, J.; O’meara, C.; Duffy, P.; Rizos, D.; Wade, M.; Donovan, A.; Boland, M.; Lonergan, P.; Evans, A. Differences between Belclare and Suffolk Ewes in Fertilization Rate, Embryo Quality and Accessory Sperm Number after Cervical or Laparoscopic Artificial Insemination. Theriogenology 2005, 63, 1995–2005. [Google Scholar] [CrossRef]
- Bugge, T.H.; Antalis, T.M.; Wu, Q. Type II Transmembrane Serine Proteases. J. Biol. Chem. 2009, 284, 23177–23181. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.S.; Zoratti, G.L.; Murray, A.S.; Bergum, C.; Tanabe, L.M.; List, K. HATL5: A Cell Surface Serine Protease Differentially Expressed in Epithelial Cancers. PLoS ONE 2014, 9, e87675. [Google Scholar] [CrossRef]
- Sales, K.U.; Friis, S.; Abusleme, L.; Moutsopoulos, N.M.; Bugge, T.H. Matriptase Promotes Inflammatory Cell Accumulation and Progresssion of Estabilished Epidermal Tumors. Oncogene 2015, 34, 4664–4672. [Google Scholar] [CrossRef] [Green Version]
- Gusmão, A.L. State-of-the-Art in the Transcervical Embryo Collection in Goats and Sheep. Acta Sci. Vet. 2011, 39, 37–42. [Google Scholar]
- Bingle, C.D.; Seal, R.L.; Craven, C.J. Systematic Nomenclature for the PLUNC/PSP/BSP30/SMGB Proteins as a Subfamily of the BPI Fold-Containing Superfamily. Biochem. Soc. Trans. 2011, 39, 977–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, T.T.; Haigh, B.J.; Broadhurst, M.K.; Hood, K.A.; Maqbool, N.J. The BPI-like/PLUNC Family Proteins in Cattle. Biochem. Soc. Trans. 2011, 39, 1006–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Burrows, J.; Card, G.L.; Attwood, G.; Wheeler, T.T.; Arcus, V.L. The Three Dimensional Structure of Bovine Salivary Protein 30b (BSP30b) and Its Interaction with Specific Rumen Bacteria. PLoS ONE 2019, 14, e0206709. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.I.; Suh, J.G.; Lee, J.W.; Hong, S.H.; Kang, T.C.; Oh, Y.S.; Ryoo, Z.Y. The Circling Mouse (Cir/Cir-C57BL/6) Has a 40-Kilobase Genomic Deletion That Includes the Transmembrane Inner Ear (Tmie) Gene. Comp. Med. 2006, 56, 476–481. [Google Scholar]
- Shin, M.J.; Lee, J.H.; Yu, D.H.; Kim, H.J.; Bae, K.B.; Yuh, H.S.; Kim, M.O.; Hyun, B.H.; Lee, S.; Park, R.; et al. Spatiotemporal Expression of Tmie in the Inner Ear of Rats during Postnatal Development. Comp. Med. 2010, 60, 288–294. [Google Scholar]
- Gleason, M.R.; Nagiel, A.; Jamet, S.; Vologodskaia, M.; Lopez-Schier, H.; Hudspeth, A.J. The Transmembrane Inner Ear (Tmie) Protein Is Essential for Normal Hearing and Balance in the Zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 21347–21352. [Google Scholar] [CrossRef] [Green Version]
- Takehiro, H.; Yasushi, H.; Yamato, F.; Mona, G.; Tetsuaki, K.; Shizu, A.; Tomoyuki, H.; Shun, A.; Mitsunori, M.; Hirofumi, H.; et al. Differential Roles of Uterine Epithelial and Stromal STAT3 Coordinate Uterine Receptivity and Embryo Attachment. Sci. Rep. 2020, 10, 15523. [Google Scholar] [CrossRef]
- Skinner, M.K.; Schmidt, M.; Savenkova, M.I.; Sadler-Riggleman, I.; Nilsson, E.E. Regulation of Granulosa and Theca Cell Transcriptomes during Ovarian Antral Follicle Development. Mol. Reprod. Dev. 2008, 75, 1457–1472. [Google Scholar] [CrossRef] [Green Version]
- Kenney, N.J.; Adkins, H.B.; Sanicola, M. Nodal and Cripto-1: Embryonic Pattern Formation Genes Involved in Mammary Gland Development and Tumorigenesis. Mammary Gland. Biol. Neoplasia 2004, 9, 133–144. [Google Scholar] [CrossRef]
- Strizzi, L.; Bianco, C.; Normanno, N.; Salomon, D. Cripto-1: A Multifunctional Modulator during Embryogenesis and Oncogenesis. Oncogene 2005, 24, 5731–5741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gershon, E.; Hadas, R.; Elbaz, M.; Booker, E.; Muchnik, M.; Kleinjan-Elazary, A.; Gray, P.C. Identification of Trophectoderm-Derived Cripto as an Essential Mediator of Embryo Implantation. Endocrinology 2018, 159, 1793–1807. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Zhong, L.; Zhang, S.; Mu, H.; Xiang, J.; Yue, L.; Han, J. Bovine Lineage Specification Revealed by Single-Cell Gene Expression Analysis from Zygote to Blastocyst. Biol. Reprod. 2007, 97, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, P.C.; Vale, W. Cripto/GRP78 Modulation of the TGF-b Pathway in Development and Oncogenesis. FEBS Lett. 2012, 586, 1836–1845. [Google Scholar]
- Bianco, C.; Rangel, M.C.; Castro, N.P.; Nagaoka, T.; Rollman, K.; Gonzales, M.; Salomon, D.S. Role of Cripto-1 in Stem Cell Maintenance and Malignant Progression. Am. J. Pathol. 2010, 177, 532–540. [Google Scholar] [CrossRef]
- Christ, A.; Herzog, K.; Willnow, T.E. LRP2, an Auxiliary Receptor That Controls Sonic Hedgehog Signaling in Development and Disease. Dev. Dyn. 2016, 245, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Seeman, P.; Van Tol, H.H. Dopamine Receptor Pharmacology. Trends. Pharmac. Sci. 1994, 15, 264–270. [Google Scholar] [CrossRef]
- Melis, M.R.; Stancampiano, R.; Argiolas, A. Hippocampal Oxytocin Mediates Apomorphine-Induced Penile Erection and Yawning. Pharmacol. Biochem. Behav. 1992, 42, 61–66. [Google Scholar] [CrossRef]
- Hirst, J.J.; Haluska, G.J.; Cook, M.J.; Novy, M.J. Plasma Oxytocin and Nocturnal Uterine Activity: Maternal but Not Fetal Concentrations Increase Progressively during Late Pregnancy and Delivery in Rhesus Monkeys. Am. J. Obstet. Gynecol. 1993, 169, 415–422. [Google Scholar] [CrossRef]
- Lyukmanova, N.E.; Shulepko, M.A.; Kudryavtsev, D.; Bychkov, M.L.; Kulbatskii, D.S.; Kasheverov, I.E.; Astapova, M.V.; Feofanov, A.V.; Thomsen, M.S.; Mikkelsen, J.D.; et al. Human Secreted Ly-6/UPAR Related Protein-1 (SLURP-1) Is a Selective Allosteric Antagonist of Alpha7 Nicotinic Acetylcholine Receptor. PLoS ONE 2016, 11, e0149733. [Google Scholar] [CrossRef] [Green Version]
- Leppert, P.C. Cervical Softening, Effacement, and Dilatation: A Complex Biochemical Cascade. J. Matern.-Fetal Neonatal Med. 1992, 1, 213–223. [Google Scholar]
- Chwalisz, K.; Benson, M.; Scholz, P.; Daum, J.; Beier, M.; Hegele-Hartung, C. Cervical Ripening with the Cytokines Interleukin 8, Interleukin 1β and Tumour Necrosis Factor α in Guinea-Pigs. Hum. Reprod. 1994, 9, 2173–2181. [Google Scholar] [CrossRef]
- Falchi, L.; Scaramuzzi, R.J. An in Vitro Investigation of the Ac- Tions of Reproductive Hormones on the Cervix of the Ewe in the Fol- Licular Stage: The Effects of 17β-estradiol, Oxytocin, FSH, and Arachi- Donic Acid on the Cervical Pathway for the Synthesis of Prostaglandin E2. Theriogenology 2015, 83, 1007–1014. [Google Scholar]
- Koets, A.P.; de Schwartz, N.; Tooten, P.; Kankofer, M.; Broekhuijsen-Davies, J.M.; Rutten, V.P.M.G.; van Leengoed, L.A.M.G.; Taverne, M.A.M.; Gruys, E. Release of Proinflammatory Cytokines Related to Luteolysis and the Periparturient Acute Phase Response in Prostaglandin-Induced Parturition in Cows. Theriogenology 1998, 49, 797–812. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Yang, D. ADAM7 Promotes the Proliferation and Invasion in Trophoblast Cells. Exp. Mol. Pathol. 2021, 121, 1046–1059. [Google Scholar] [CrossRef]
- De Rensis, F.; Valentini, R.; Gorrieri, F.; Bottarelli, E.; Lopez-Gatius, F. Inducing Ovulation with HCG Improves the Fertility of Dairy Cows during the Warm Season. Theriogenology 2008, 69, 1077–1082. [Google Scholar] [CrossRef]
- Kalluri, R. Basement Membranes: Structure, Assembly and Role in Tumour Angiogenesis. Nat. Rev. Cancer 2003, 3, 422–433. [Google Scholar]
- Kershaw-Young, C.M.; Khalid, M.; McGowan, M.R.; Pitsillides, A.A.; Scaramuzzi, R.J. The MRNA Expression of Prostaglandin E Receptors EP2 and EP4 and the Changes in Glycosaminoglycans in the Sheep Cervix during the Estrous Cycle. Theriogenology 2009, 72, 251–261. [Google Scholar] [CrossRef]
- Kershaw, C.M.; Scaramuzzi, R.J.; McGowan, M.R.; Wheeler-Jones, C.P.; Khalid, M. The Expression of Prostaglandin Endoperoxide Synthase 2 Messenger RNA and the Proportion of Smooth Muscle and Collagen in the Sheep Cervix during the Estrous Cycle. Biol. Reprod. 2007, 76, 124–129. [Google Scholar]
- Rodríguez-Piñón, M.; Tasende, C.; Casuriaga, D.; Bielli, A.; Genovese, P.; Garófalo, E.G. Collagen and Matrix Metalloproteinase-2 and -9 in the Ewe Cérvix during the Estrous Cycle. Theriogenology 2015, 84, 818–826. [Google Scholar] [CrossRef]
- Wellik, D.M. Hox Genes and Vertebrate Axial Pattern. Curr. Top. Dev. Biol. 2009, 88, 257–278. [Google Scholar] [PubMed]
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox Genes and Regional Patterning of the Vertebrate Body Plan. Dev. Biol. 2010, 244, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Fernàndez, J. Hox, ParaHox, ProtoHox: Facts and Guesses. Heredity (Edinb) 2005, 94, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Davenne, M.; Maconochie, M.K.; Neun, R.; Pattyn, A.; Chambon, P.; Krumlauf, P.; Rijli, F.M. Hoxa2 and Hoxb2 Control Dorsoventral Patterns of Neuronal Development in the Rostral Hindbrain. Neuron 1999, 4, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Capecchi, M.R. Paralogous Mouse Hox Genes, Hoxa9, Hoxb9, and Hoxd9, Function Together to Control Development of the Mammary Gland in Response to Pregnancy. Proc. Natl. Acad. Sci. USA 1999, 96, 541–546. [Google Scholar] [CrossRef] [Green Version]
- Sakiyama, J.; Yokouchi, Y.; Kuroiwa, A. Coordinated Expression of Hoxb Genes and Signaling Molecules during Development of the Chick Respiratory Tract. Dev. Biol. 2000, 227, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Sha, S.; Gu, Y.; Xu, B.; Hu, H.; Yang, Y.; Kong, X.; Wu, K. Decreased Expression of HOXB9 Is Related to Poor Overall Survival in Patients with Gastric Carcinoma. Dig. Liver Dis. 2013, 45, 422–429. [Google Scholar] [CrossRef]
- Zhan, J.; Wang, P.; Niu, M.; Wang, Y.; Zhu, X.; Guo, Y.; Zhang, H. High Expression of Transcriptional Factor HoxB9 Predicts Poor Prognosis in Patients with Lung Adenocarcinoma. Histopathology 2015, 66, 955–965. [Google Scholar] [CrossRef]
- Perkins, A.C.; Cory, C. Conditional Immortalization of Mouse Myelomonocytic, Megakaryocytic and Mast Cell Progenitors by the Hox-2.4 Homeobox Gene. EMBO J. 1993, 12, 3835–3846. [Google Scholar] [CrossRef]
- Van Deren, D.A.; De, S.; Xu, B.; Eschenbacher, K.M.; Zhang, S.; Capecchi, M.R. Defining the Hoxb8 Cell Lineage during Murine Definitive Hematopoiesis. Development 2022, 15, 49. [Google Scholar] [CrossRef]
- De las Heras-Saldana, S.; Chung, K.Y.; Lee, S.H.; Gondro, C. Gene Expression of Hanwoo Satellite Cell Differentiation in Longissimus Dorsi and Semimembranosus. BMC Genom. 2019, 20, 156. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Herrera, A.L.; Salgado-Bernabe, M.; Velazquez-Velazquez, C.K.; Salcedo-Vargas, M.; Andrade-Manzano, A.; Avila-Moreno, F.; Pina-Sanchez, P. Increased Expression of HOXB2 and HOXB13 Proteins Is Associated with HPV Infection and Cervical Cancer Progression. Asian Pac. J. Cancer Prev. 2015, 16, 1349–1353. [Google Scholar] [CrossRef] [Green Version]
- Houghton, L.; Rosenthal, N. Regulation of a Muscle-Specific Transgene by Persistent Expression of Hox Genes in Postnatal Murine Limb Muscle. Dev. Dyn. 1999, 216, 385–397. [Google Scholar] [CrossRef]
- Davis, R.J. Signal Transduction by the JNK Group of MAP Kinases. Cell 2000, 103, 239–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, S.R.; Brunet, A.; Greenberg, M.E. Cellular Survival: A Play in Three Akts. Genes Dev. 1999, 13, 2905–2927. [Google Scholar] [CrossRef] [PubMed]
- Kohn, A.D.; Kovacina, K.S.; Roth, R.A. Insulin Stimulates the Kinase Activity of RAC-PK, a Pleckstrin Homology Domain Containing Ser/Thr Kinase. EMBO J. 1995, 14, 4288–4295. [Google Scholar] [CrossRef]
- Kohn, A.D.; Summers, S.A.; Birnbaum, M.J.; Roth, R.A. Expression of a Constitutively Active Akt Ser/Thr Kinase in 3T3-L1 Adipocytes Stimulates Glucose Uptake and Glucose Transporter 4 Translocation. J. Biol. Chem. 1996, 271, 31372–31378. [Google Scholar] [CrossRef] [Green Version]
- Lue, H.; Thiele, M.; Franz, J.; Dahl, E.; Speckgens, S.; Leng, L.; Fingerle-Rowson, G.; Bucala, R.; Luscher, B.; Bernhagen, J. Macrophage Migration Inhibitory Factor (MIF) Promotes Cell Survival by Activation of the Akt Pathway and Role for CSN5/JAB1 in the Control of Autocrine MIF Activity. Oncogene 2007, 26, 5046–5059. [Google Scholar] [CrossRef] [Green Version]
- Manning, B.D.; Cantley, L.C. AKT/PKB Signaling: Navigating Downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [Green Version]
- Brazil, D.P.; Hemmings, B.A. Ten Years of Protein Kinase B Signalling: A Hard Akt to Follow. Trends Biochem. Sci. 2001, 26, 657–664. [Google Scholar] [CrossRef]
- Bruhn, M.A.; Pearson, R.B.; Hannan, R.D.; Sheppard, K.E. AKT-Independent PI3-K Signaling in Cancer—Emerging Role for SGK3. Cancer Manag. Res. 2013, 5, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fayard, E.; Tintignac, L.A.; Baudry, A.; Hemmings, B.A. Protein Kinase B/Akt at a Glance. J. Cell Sci. 2005, 118, 5675–5678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willems, L.; Tamburini, J.; Chapuis, N.; Lacombe, C.; Mayeux, P.; Bouscary, D. PI3K and MTOR Signaling Pathways in Cancer: New Data on Targeted Therapies. Curr. Oncol. Rep. 2012, 14, 129–138. [Google Scholar] [CrossRef]
- Geengard, P. The Neurobiology of Slow Sinaptic Transmission. Science (1979) 2001, 294, 1024–1030. [Google Scholar]
- Duan, J.; Yu, Y.; Li, Y.; Li, Y.; Liu, H.; Jing, L.; Yang, M.; Wang, J.; Li, C.; Sun, Z. Low-Dose Exposure of Silica Nanoparticles Induces Cardiac Dysfunction via Neutrophil-Mediated Inflammation and Cardiac Contraction in Zebrafish Embryos. Nanotoxicology 2016, 10, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Dhirachaikulpanich, D.; Li, X.; Porter, L.F.; Paraoan, L. Integrated Microarray and RNAseq Transcriptomic Analysis of Retinal Pigment Epithelium/Choroid in Age-Related Macular Degeneration. Front. Cell Dev. Biol. 2020, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ramıírez, A.R.; Castro, M.A.; Ramio, C.A.L.; Rivera, M.M.; Torres, M.; Rigau, T.; RodrÍguez-Gil, J.E.; Concha, I.I. The Presence and Function of Dopamine Type 2 Receptors in Boar Sperm: A Possible Role for Dopamine in Viability, Capacitation, and Modulation of Sperm Motility1. Biol. Reprod. 2009, 80, 753–761. [Google Scholar] [CrossRef]
- Fahy, J.V.; Dickey, B.F. Airway Mucus Function and Dysfunction. N. Engl. J. Med. 2010, 363, 2233–2247. [Google Scholar] [CrossRef] [Green Version]
- Eriksen, G.V.; Carlstedt, I.; Uldbjerg, N.; Ernst, E. Cervical Mucins Affect the Motility of Human Spermatozoa in Vitro. Fertil. Steril. 1998, 70, 350–354. [Google Scholar] [CrossRef]
- Brayman, M.; Thathiah, A.; Carson, D.D. MUC1: A Multifunctional Cell Surface Component of Reproductive Tissue Epithelia. Reprod. Biol. Endocrinol. 2004, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- McAuley, J.; Linden, S.K.; Png, C.W.; King, R.M.; Pennington, H.L.; Gendler, S.J.; Florin, T.H.; Hill, G.; Korolik, V.; McGuckin, M.A. MUC1 Cell Surface Mucin Is a Critical Element of the Mucosal Barrier to Infection. J. Clin. Investig. 2007, 117, 2313–2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osteen, K.G.; Rodgers, W.H.; Gaire, M.; Hargrove, J.T.; Gorstein, F.; Matrisian, L.M. Stromal511 Epithelial Interaction Mediates Steroidal Regulation of Metalloproteinase Expression in 512 Human Endometrium. Proc. Natl. Acad. Sci. USA 1994, 91, 10129–10133. [Google Scholar] [CrossRef] [Green Version]
- Bairagi, S.; Grazul-Bilska, A.T.; Borowicz, P.P.; Reyaz, A.; Valkov, V.; Reynolds, L.P. Placental Development during Early Pregnancy in Sheep: Progesterone and Estrogen Receptor Protein Expression. Theriogenology 2018, 114, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Elsbach, P.; Olsson, I.; Odeberg, H. Purification and Characterization of a Potent Bactericidal and Membrane Active Protein from the Granules of Human Polymorphonuclear Leukocytes. J. Biol. Chem. 1978, 253, 2664–2672. [Google Scholar] [CrossRef] [PubMed]
- Elsbach, P. The Bactericidal/Permeability-Increasing Protein (BPI) in Antibacterial Host Defense. J. Leukoc. Biol. 1998, 64, 14–18. [Google Scholar] [CrossRef]
- Hou, D.; Zhou, X.; Zhong, X.; Settles, M.L.; Herring, J.; Wang, L.; Abdo, Z.; Forney, L.J.; Xu, C. Microbiota of the Seminal Fluid from Healthy and Infertile Men. Fertil. Steril. 2013, 100, 1261–1269.e3. [Google Scholar] [CrossRef] [Green Version]
- Takeda, Y.; Demura, M.; Yoneda, T.; Takeda, Y. DNA Methylation of the Angiotensinogen Gene, AGT, and the Aldosterone Synthase Gene, CYP11B2 in Cardiovascular Diseases. Int. J. Mol. Sci. 2021, 22, 4587. [Google Scholar] [CrossRef]
- Kool, M.J.; Van De Bree, J.E.; Bodde, H.E.; Elgersma, Y.; Van Woerden, G.M. The Molecular, Temporal and Region-Specific Requirements of the Beta Isoform of Calcium/Calmodulin-Dependent Protein Kinase Type 2 (CAMK2B) in Mouse Locomotion. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Bird, I.M.; Chen, D.B.; Magness, R.R. Angiotensin II Regulation of Ovine Fetoplacental Artery Endothelial Functions: Interactions with Nitric Oxide. J. Physiol. 2005, 565, 59–69. [Google Scholar] [CrossRef]
- Brosnihan, K.B.; Neves, L.A.A.; Anton, L.; Joyner, J.; Valdes, G.; Merril, D.C. Enhanced Expression of Ang-(1-7) during Pregnancy. Braz. J. Med. Biol. Res. 2004, 37, 1255–1262. [Google Scholar] [CrossRef] [Green Version]
- Costa, A.S.; Junior, A.S.; Viana, G.E.N.; Muratori, M.C.S.; Costa, A.P.R. Inhibition of Angiotensin-Converting Enzyme Increases Oestradiol Production in Ewes Submitted to Oestrous Synchronization Protocol. Reprod. Domest. Anim. 2014, 49, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Giani, J.F.; Gironacci, M.M.; Muñoz, M.C.; Peña, C.; Turun, D.; Dominici, F.P. Angiotensin-(1 7) Stimulates the Phosphorylation of JAK2, IRS-1 and Akt in Rat Heart in Vivo: Role of the AT1 and Mas Receptors. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, 1154–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampaio, W.O.; Souza Dos Santos, R.A.; Faria-Silva, R.; Da Mata Machado, L.T.; Schiffrin, E.L.; Touyz, R.M. Angiotensin-(1–7) through Receptor Mas Mediates Endothelial Nitric Oxide Synthase Activation via Akt-Dependent Pathways. Hypertension 2007, 49, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, J.D.; Ferraz, J.B.S.; Meirelles, F.V.; Nociti, R.P.; Oliveira, M.E.F. An Exploratory Data Analysis from Ovine and Bovine RNA-Seq Identifies Pathways and Key Genes Related to Cervical Dilatation. Animals 2023, 13, 2052. https://doi.org/10.3390/ani13132052
Gonçalves JD, Ferraz JBS, Meirelles FV, Nociti RP, Oliveira MEF. An Exploratory Data Analysis from Ovine and Bovine RNA-Seq Identifies Pathways and Key Genes Related to Cervical Dilatation. Animals. 2023; 13(13):2052. https://doi.org/10.3390/ani13132052
Chicago/Turabian StyleGonçalves, Joedson Dantas, José Bento Sterman Ferraz, Flávio Vieira Meirelles, Ricardo Perecin Nociti, and Maria Emilia Franco Oliveira. 2023. "An Exploratory Data Analysis from Ovine and Bovine RNA-Seq Identifies Pathways and Key Genes Related to Cervical Dilatation" Animals 13, no. 13: 2052. https://doi.org/10.3390/ani13132052
APA StyleGonçalves, J. D., Ferraz, J. B. S., Meirelles, F. V., Nociti, R. P., & Oliveira, M. E. F. (2023). An Exploratory Data Analysis from Ovine and Bovine RNA-Seq Identifies Pathways and Key Genes Related to Cervical Dilatation. Animals, 13(13), 2052. https://doi.org/10.3390/ani13132052






