How Does Nutrition Affect the Epigenetic Changes in Dairy Cows?
Abstract
Simple Summary
Abstract
1. Introduction
2. Nutrition in Dairy Cows
3. Epigenetic Regulators
3.1. DNA Methylation
3.2. Histone Modifications
3.3. Small Non-Coding RNA: miRNAs as Epigenetic Regulators
4. Epigenetic Regulation and Nutrition
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OECD; FAO. OECD-FAO Agricultural Outlook 2022–2031. In OECD-FAO Agricultural Outlook; OECD: Paris, France, 2022; ISBN 978-92-64-58870-7. [Google Scholar]
- Adesogan, A.T.; Dahl, G.E. MILK Symposium Introduction: Dairy Production in Developing Countries. J. Dairy Sci. 2020, 103, 9677–9680. [Google Scholar] [CrossRef] [PubMed]
- Dror, D.K.; Allen, L.H. Overview of Nutrients in Human Milk. Adv. Nutr. 2018, 9, 278S–294S. [Google Scholar] [CrossRef] [PubMed]
- Górska-Warsewicz, H.; Rejman, K.; Laskowski, W.; Czeczotko, M. Milk and Dairy Products and Their Nutritional Contribution to the Average Polish Diet. Nutrients 2019, 11, 1771. [Google Scholar] [CrossRef] [PubMed]
- Fekete, Á.A.; Givens, D.I.; Lovegrove, J.A. The Impact of Milk Proteins and Peptides on Blood Pressure and Vascular Function: A Review of Evidence from Human Intervention Studies. Nutr. Res. Rev. 2013, 26, 177–190. [Google Scholar] [CrossRef]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef]
- Rizzoli, R. Dairy Products, Yogurts, and Bone Health. Am. J. Clin. Nutr. 2014, 99, 1256S–1262S. [Google Scholar] [CrossRef]
- Baumgard, L.H.; Collier, R.J.; Bauman, D.E. A 100-Year Review: Regulation of Nutrient Partitioning to Support Lactation. J. Dairy Sci. 2017, 100, 10353–10366. [Google Scholar] [CrossRef]
- Bewley, J.M.; Robertson, L.M.; Eckelkamp, E.A. A 100-Year Review: Lactating Dairy Cattle Housing Management. J. Dairy Sci. 2017, 100, 10418–10431. [Google Scholar] [CrossRef]
- Sehested, J.; Gaillard, C.; Lehmann, J.O.; Maciel, G.M.; Vestergaard, M.; Weisbjerg, M.R.; Mogensen, L.; Larsen, L.B.; Poulsen, N.A.; Kristensen, T. Review: Extended Lactation in Dairy Cattle. Animal 2019, 13, s65–s74. [Google Scholar] [CrossRef]
- Hagan, B.A.; Moro-Mendez, J.; Cue, R.I. Realized Genetic Selection Differentials in Canadian Holstein Dairy Herds. J. Dairy Sci. 2020, 103, 1651–1666. [Google Scholar] [CrossRef]
- Jaton, C.; Schenkel, F.S.; Chud, T.C.S.; Malchiodi, F.; Sargolzaei, M.; Price, C.A.; Canovàs, A.; Baes, C.; Miglior, F. Genetic and Genomic Analyses of Embryo Production in Dairy Cattle. Reprod. Fertil. Dev. 2019, 32, 50–55. [Google Scholar] [CrossRef]
- Martin, P.; Barkema, H.W.; Brito, L.F.; Narayana, S.G.; Miglior, F. Symposium Review: Novel Strategies to Genetically Improve Mastitis Resistance in Dairy Cattle. J. Dairy Sci. 2018, 101, 2724–2736. [Google Scholar] [CrossRef]
- Sun, H.Z.; Plastow, G.; Guan, L.L. Invited Review: Advances and Challenges in Application of Feedomics to Improve Dairy Cow Production and Health. J. Dairy Sci. 2019, 102, 5853–5870. [Google Scholar] [CrossRef]
- Fodor, I.; Abonyi-Tóth, Z.; Ózsvári, L. Management Practices Associated with Reproductive Performance in Holstein Cows on Large Commercial Dairy Farms. Animal 2018, 12, 2401–2406. [Google Scholar] [CrossRef]
- Lacasse, P.; Vanacker, N.; Ollier, S.; Ster, C. Innovative Dairy Cow Management to Improve Resistance to Metabolic and Infectious Diseases during the Transition Period. Res. Vet. Sci. 2018, 116, 40–46. [Google Scholar] [CrossRef]
- McMullen, C.K.; Sargeant, J.M.; Kelton, D.F.; Churchill, K.J.; Cousins, K.S.; Winder, C.B. Modifiable Management Practices to Improve Udder Health in Dairy Cattle during the Dry Period and Early Lactation: A Scoping Review. J. Dairy Sci. 2021, 104, 10143–10157. [Google Scholar] [CrossRef]
- Erickson, P.S.; Kalscheur, K.F. Nutrition and Feeding of Dairy Cattle. Anim. Agric. 2020, 157–180. [Google Scholar] [CrossRef]
- Hristov, A.N.; Melgar, A.; Wasson, D.; Arndt, C. Symposium Review: Effective Nutritional Strategies to Mitigate Enteric Methane in Dairy Cattle. J. Dairy Sci. 2022, 105, 8543–8557. [Google Scholar] [CrossRef]
- Jirtle, R.L.; Skinner, M.K. Environmental Epigenomics and Disease Susceptibility. Nat. Rev. Genet. 2007, 8, 253–262. [Google Scholar] [CrossRef]
- Marín-García, P.J.; Llobat, L. How Does Protein Nutrition Affect the Epigenetic Changes in Pig? A Review. Animals 2021, 11, 544. [Google Scholar] [CrossRef]
- Amanzadeh, J.; Gitomer, W.L.; Zerwekh, J.E.; Preisig, P.A.; Moe, O.W.; Pak, C.Y.C.; Levi, M. Effect of High Protein Diet on Stone-Forming Propensity and Bone Loss in Rats. Kidney Int. 2003, 64, 2142–2149. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Nordgren, K.K.S.; Chai, Y.; Hebbring, S.J.; Jenkins, G.D.; Abo, R.P.; Peng, Y.; Pelleymounter, L.L.; Moon, I.; Eckloff, B.W.; et al. Human Liver Methionine Cycle: MAT1A and GNMT Gene Resequencing, Functional Genomics, and Hepatic Genotype-Phenotype Correlation. Drug Metab. Dispos. 2012, 40, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Palmquist, D.L.; Beaulieu, A.D.; Barbano, D.M. Feed and Animal Factors Influencing Milk Fat Composition. J. Dairy Sci. 1993, 76, 1753–1771. [Google Scholar] [CrossRef]
- Castro, M.M.D.; Albino, R.L.; Rodrigues, J.P.P.; Sguizzato, A.L.L.; Santos, M.M.F.; Rotta, P.P.; Caton, J.S.; Moraes, L.E.F.D.; Silva, F.F.; Marcondes, M.I. Energy and Protein Requirements of Holstein × Gyr Crossbred Heifers. Animal 2020, 14, 1857–1866. [Google Scholar] [CrossRef]
- Marcondes, M.I.; Silva, A.L. Determination of Energy and Protein Requirements of Preweaned Dairy Calves: A Multistudy Approach. J. Dairy Sci. 2021, 104, 11553–11566. [Google Scholar] [CrossRef] [PubMed]
- Salah, N.; Sauvant, D.; Archimède, H. Nutritional Requirements of Sheep, Goats and Cattle in Warm Climates: A Meta-Analysis. Animal 2014, 8, 1439–1447. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Dairy Cattle: Seventh Revised Edition; National Academies Press: Washington, DC, USA, 2001; ISBN 978-0-309-06997-7.
- Duplessis, M.; Pellerin, D.; Robichaud, R.; Fadul-Pacheco, L.; Girard, C.L. Impact of Diet Management and Composition on Vitamin B12 Concentration in Milk of Holstein Cows. Animal 2019, 13, 2101–2109. [Google Scholar] [CrossRef]
- Vicente, F.; Santiago, C.; Jiménez-Calderón, J.D.; Martínez-Fernández, A. Capacity of Milk Composition to Identify the Feeding System Used to Feed Dairy Cows. J. Dairy Res. 2017, 84, 254–263. [Google Scholar] [CrossRef]
- Foroutan, A.; Guo, A.C.; Vazquez-Fresno, R.; Lipfert, M.; Zhang, L.; Zheng, J.; Badran, H.; Budinski, Z.; Mandal, R.; Ametaj, B.N.; et al. Chemical Composition of Commercial Cow’s Milk. J. Agric. Food Chem. 2019, 67, 4897–4914. [Google Scholar] [CrossRef]
- Cant, J.P.; McBride, B.W. Mathematical Analysis of the Relationship between Blood Flow and Uptake of Nutrients in the Mammary Glands of a Lactating Cow. J. Dairy Res. 1995, 62, 405–422. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The Rumen Microbiome: A Crucial Consideration When Optimising Milk and Meat Production and Nitrogen Utilisation Efficiency. Gut. Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Schwab, C.G.; Broderick, G.A. A 100-Year Review: Protein and Amino Acid Nutrition in Dairy Cows. J. Dairy Sci. 2017, 100, 10094–10112. [Google Scholar] [CrossRef]
- PDIN INRA 2007. Tables of Composition and Nutritional Values of Feed Materials INRA CIRAD AFZ. Available online: https://www.feedtables.com/content/pdin-inra-2007 (accessed on 4 May 2023).
- Smith, Z.D.; Meissner, A. DNA Methylation: Roles in Mammalian Development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Hoang, N.M.; Rui, L. DNA Methyltransferases in Hematological Malignancies. J. Genet. Genom. 2020, 47, 361–372. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for de Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef]
- Hamidi, T.; Singh, A.K.; Chen, T. Genetic Alterations of DNA Methylation Machinery in Human Diseases. Epigenomics 2015, 7, 247–265. [Google Scholar] [CrossRef]
- Deng, J.; Szyf, M. Multiple Isoforms of DNA Methyltransferase Are Encoded by the Vertebrate Cytosine DNA Methyltransferase Gene. J. Biol. Chem. 1998, 273, 22869–22872. [Google Scholar] [CrossRef]
- Leonhardt, H.; Bestor, T.H. Structure, Function and Regulation of Mammalian DNA Methyltransferase. In DNA Methylation: Molecular Biology and Biological Significance; Jost, J.-P., Saluz, H.-P., Eds.; Birkhäuser: Basel, Switzerland, 1993; pp. 109–119. ISBN 978-3-0348-9118-9. [Google Scholar]
- Robertson, K.D.; Uzvolgyi, E.; Liang, G.; Talmadge, C.; Sumegi, J.; Gonzales, F.A.; Jones, P.A. The Human DNA Methyltransferases (DNMTs) 1, 3a and 3b: Coordinate MRNA Expression in Normal Tissues and Overexpression in Tumors. Nucleic Acids Res. 1999, 27, 2291–2298. [Google Scholar] [CrossRef]
- Montaner-Angoiti, E.; Marín-García, P.J.; Llobat, L. Epigenetic Alterations in Canine Malignant Lymphoma: Future and Clinical Outcomes. Animals 2023, 13, 468. [Google Scholar] [CrossRef]
- Luu, H.N.; Wang, R.; Jin, A.; Koh, W.-P.; Yuan, J.-M. The Association Between Dietary Vitamin B12 and Lung Cancer Risk: Findings from a Prospective Cohort Study. Eur. J. Cancer Prev. 2021, 30, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.; Matte, A.; Perfilyev, A.; de Mello, V.D.; Käkelä, P.; Pihlajamäki, J.; Ling, C. Epigenetic Alterations in Human Liver From Subjects With Type 2 Diabetes in Parallel With Reduced Folate Levels. J. Clin. Endocrinol. Metab. 2015, 100, E1491–E1501. [Google Scholar] [CrossRef] [PubMed]
- Salbaum, J.M.; Kappen, C. Genetic and Epigenomic Footprints of Folate. Prog. Mol. Biol. Transl. Sci. 2012, 108, 129–158. [Google Scholar] [CrossRef]
- Zeisel, S. Choline, Other Methyl-Donors and Epigenetics. Nutrients 2017, 9, 445. [Google Scholar] [CrossRef] [PubMed]
- Kovacheva, V.P.; Mellott, T.J.; Davison, J.M.; Wagner, N.; Lopez-Coviella, I.; Schnitzler, A.C.; Blusztajn, J.K. Gestational Choline Deficiency Causes Global and Igf2 Gene DNA Hypermethylation by Up-Regulation of Dnmt1 Expression. J. Biol. Chem. 2007, 282, 31777–31788. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent Epigenetic Differences Associated with Prenatal Exposure to Famine in Humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef]
- Murdoch, B.M.; Murdoch, G.K.; Greenwood, S.; McKay, S. Nutritional Influence on Epigenetic Marks and Effect on Livestock Production. Front. Genet. 2016, 7, 182. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, Q.; Li, X.; Wang, M.; Cai, D.; Li, X.; Zhao, R. In Ovo Injection of Betaine Affects Hepatic Cholesterol Metabolism through Epigenetic Gene Regulation in Newly Hatched Chicks. PLoS ONE 2015, 10, e0122643. [Google Scholar] [CrossRef]
- Cai, D.; Jia, Y.; Song, H.; Sui, S.; Lu, J.; Jiang, Z.; Zhao, R. Betaine Supplementation in Maternal Diet Modulates the Epigenetic Regulation of Hepatic Gluconeogenic Genes in Neonatal Piglets. PLoS ONE 2014, 9, e105504. [Google Scholar] [CrossRef]
- Fan, Y.; Vilgalys, T.P.; Sun, S.; Peng, Q.; Tung, J.; Zhou, X. IMAGE: High-Powered Detection of Genetic Effects on DNA Methylation Using Integrated Methylation QTL Mapping and Allele-Specific Analysis. Genome Biol. 2019, 20, 220. [Google Scholar] [CrossRef]
- Gibbs, J.R.; van der Brug, M.P.; Hernandez, D.G.; Traynor, B.J.; Nalls, M.A.; Lai, S.-L.; Arepalli, S.; Dillman, A.; Rafferty, I.P.; Troncoso, J.; et al. Abundant Quantitative Trait Loci Exist for DNA Methylation and Gene Expression in Human Brain. PLoS Genet 2010, 6, e1000952. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell. Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Esmaeili, M.; Blythe, S.A.; Tobias, J.W.; Zhang, K.; Yang, J.; Klein, P.S. Chromatin Accessibility and Histone Acetylation in the Regulation of Competence in Early Development. Dev. Biol. 2020, 462, 20–35. [Google Scholar] [CrossRef]
- Izzo, A.; Schneider, R. The Role of Linker Histone H1 Modifications in the Regulation of Gene Expression and Chromatin Dynamics. Biochim. Biophys. Acta 2016, 1859, 486–495. [Google Scholar] [CrossRef]
- Martire, S.; Banaszynski, L.A. The Roles of Histone Variants in Fine-Tuning Chromatin Organization and Function. Nat. Rev. Mol. Cell. Biol. 2020, 21, 522–541. [Google Scholar] [CrossRef]
- Xhemalce, B.; Dawson, M.; Bannister, A. Histone Modifications. In Reviews in Cell Biology and Molecular Medicine; Wiley: Hoboken, NJ, USA, 2011; ISBN 978-3-527-60090-8. [Google Scholar]
- Oki, M.; Aihara, H.; Ito, T. Role of Histone Phosphorylation in Chromatin Dynamics and Its Implications in Diseases. Subcell. Biochem. 2007, 41, 319–336. [Google Scholar]
- Bannister, A.J.; Schneider, R.; Kouzarides, T. Histone Methylation: Dynamic or Static? Cell 2002, 109, 801–806. [Google Scholar] [CrossRef]
- Copeland, R.A.; Solomon, M.E.; Richon, V.M. Protein Methyltransferases as a Target Class for Drug Discovery. Nat. Rev. Drug Discov. 2009, 8, 724–732. [Google Scholar] [CrossRef]
- Dahl, J.A.; Reiner, A.H.; Klungland, A.; Wakayama, T.; Collas, P. Histone H3 Lysine 27 Methylation Asymmetry on Developmentally-Regulated Promoters Distinguish the First Two Lineages in Mouse Preimplantation Embryos. PLoS ONE 2010, 5, e9150. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and Regulation of Histone Methylation in Animal Development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Mitani, S.; Du, H.; Hall, D.H.; Driscoll, M.; Chalfie, M. Combinatorial Control of Touch Receptor Neuron Expression in Caenorhabditis Elegans. Development 1993, 119, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-Nucleotide Let-7 RNA Regulates Developmental Timing in Caenorhabditis Elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Doench, J.G.; Sharp, P.A. Specificity of MicroRNA Target Selection in Translational Repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; Kerin, M.J. MiRNAs as Biomarkers and Therapeutic Targets in Cancer. Curr. Opin. Pharmacol. 2010, 10, 543–550. [Google Scholar] [CrossRef]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cell. Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- German, C.A.; Shapiro, M.D. Small Interfering RNA Therapeutic Inclisiran: A New Approach to Targeting PCSK9. BioDrugs 2020, 34, 1–9. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Shuwen, H.; Xi, Y.; Quan, Q.; Yin, J.; Miao, D. Can Small Nucleolar RNA Be a Novel Molecular Target for Hepatocellular Carcinoma? Gene 2020, 733, 144384. [Google Scholar] [CrossRef]
- Bravo, J.I.; Nozownik, S.; Danthi, P.S.; Benayoun, B.A. Transposable Elements, Circular RNAs and Mitochondrial Transcription in Age-Related Genomic Regulation. Development 2020, 147, dev175786. [Google Scholar] [CrossRef]
- Kumar, S.; Gonzalez, E.A.; Rameshwar, P.; Etchegaray, J.-P. Non-Coding RNAs as Mediators of Epigenetic Changes in Malignancies. Cancers 2020, 12, 3657. [Google Scholar] [CrossRef]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-Coding RNA Regulatory Networks. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Llobat, L.; Gourbault, O. Role of MicroRNAs in Human Osteosarcoma: Future Perspectives. Biomedicines 2021, 9, 463. [Google Scholar] [CrossRef]
- Xie, X.-L.; Yu, Y.; Yuan, Z.-F.; Yang, J.; Ma, P.-P.; Li, D.-C.; Yu, S.-K.; An, F.; Feng, X.-J.; Zhang, Y. Comparative analysis on content and distribution of CpG sites in milk production traits and mastitis-related genes in dairy cattle. Yi Chuan Hered. 2012, 34, 437–444. [Google Scholar] [CrossRef]
- Wang, M.; Bissonnette, N.; Dudemaine, P.-L.; Zhao, X.; Ibeagha-Awemu, E.M. Whole Genome DNA Methylation Variations in Mammary Gland Tissues from Holstein Cattle Producing Milk with Various Fat and Protein Contents. Genes 2021, 12, 1727. [Google Scholar] [CrossRef]
- Vanselow, J.; Yang, W.; Herrmann, J.; Zerbe, H.; Schuberth, H.-J.; Petzl, W.; Tomek, W.; Seyfert, H.-M. DNA-Remethylation around a STAT5-Binding Enhancer in the AlphaS1-Casein Promoter Is Associated with Abrupt Shutdown of AlphaS1-Casein Synthesis during Acute Mastitis. J. Mol. Endocrinol. 2006, 37, 463–477. [Google Scholar] [CrossRef]
- Singh, K.; Erdman, R.A.; Swanson, K.M.; Molenaar, A.J.; Maqbool, N.J.; Wheeler, T.T.; Arias, J.A.; Quinn-Walsh, E.C.; Stelwagen, K. Epigenetic Regulation of Milk Production in Dairy Cows. J. Mammary Gland Biol. Neoplasia 2010, 15, 101–112. [Google Scholar] [CrossRef]
- Singh, K.; Molenaar, A.J.; Swanson, K.M.; Gudex, B.; Arias, J.A.; Erdman, R.A.; Stelwagen, K. Epigenetics: A Possible Role in Acute and Transgenerational Regulation of Dairy Cow Milk Production. Animal 2012, 6, 375–381. [Google Scholar] [CrossRef]
- Tian, P.; Luo, Y.; Li, X.; Tian, J.; Tao, S.; Hua, C.; Geng, Y.; Ni, Y.; Zhao, R. Negative Effects of Long-Term Feeding of High-Grain Diets to Lactating Goats on Milk Fat Production and Composition by Regulating Gene Expression and DNA Methylation in the Mammary Gland. J. Anim. Sci. Biotechnol. 2017, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, J.; Zhang, Q.; Jiang, L. Regulation of DNA Methylation on EEF1D and RPL8 Expression in Cattle. Genetica 2017, 145, 387–395. [Google Scholar] [CrossRef] [PubMed]
- DelCurto, H.; Wu, G.; Satterfield, M.C. Nutrition and Reproduction: Links to Epigenetics and Metabolic Syndrome in Offspring. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine Inhibits Autophagy and Promotes Growth by Inducing the SAM-Responsive Methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Batistel, F.; Ma, Y.; Alharthi, A.S.M.; Parys, C.; Loor, J.J. Methionine Supply Alters Mammary Gland Antioxidant Gene Networks via Phosphorylation of Nuclear Factor Erythroid 2-like 2 (NFE2L2) Protein in Dairy Cows during the Periparturient Period. J. Dairy Sci. 2018, 101, 8505–8512. [Google Scholar] [CrossRef]
- Jacometo, C.B.; Alharthi, A.S.; Zhou, Z.; Luchini, D.; Loor, J.J. Maternal Supply of Methionine during Late Pregnancy Is Associated with Changes in Immune Function and Abundance of MicroRNA and MRNA in Holstein Calf Polymorphonuclear Leukocytes. J. Dairy Sci. 2018, 101, 8146–8158. [Google Scholar] [CrossRef]
- Liang, Y.; Batistel, F.; Parys, C.; Loor, J.J. Glutathione Metabolism and Nuclear Factor Erythroid 2-like 2 (NFE2L2)-Related Proteins in Adipose Tissue Are Altered by Supply of Ethyl-Cellulose Rumen-Protected Methionine in Peripartal Holstein Cows. J. Dairy Sci. 2019, 102, 5530–5541. [Google Scholar] [CrossRef]
- Zhou, Z.; Garrow, T.A.; Dong, X.; Luchini, D.N.; Loor, J.J. Hepatic Activity and Transcription of Betaine-Homocysteine Methyltransferase, Methionine Synthase, and Cystathionine Synthase in Periparturient Dairy Cows Are Altered to Different Extents by Supply of Methionine and Choline. J. Nutr. 2017, 147, 11–19. [Google Scholar] [CrossRef]
- Ma, N.; Liang, Y.; Cardoso, F.F.; Parys, C.; Cardoso, F.C.; Shen, X.; Loor, J.J. Insulin Signaling and Antioxidant Proteins in Adipose Tissue Explants from Dairy Cows Challenged with Hydrogen Peroxide Are Altered by Supplementation of Arginine or Arginine plus Methionine. J. Anim. Sci. 2022, 100, skac036. [Google Scholar] [CrossRef]
- Coleman, D.N.; Vailati-Riboni, M.; Elolimy, A.A.; Cardoso, F.C.; Rodriguez-Zas, S.L.; Miura, M.; Pan, Y.-X.; Loor, J.J. Hepatic Betaine-Homocysteine Methyltransferase and Methionine Synthase Activity and Intermediates of the Methionine Cycle Are Altered by Choline Supply during Negative Energy Balance in Holstein Cows. J. Dairy Sci. 2019, 102, 8305–8318. [Google Scholar] [CrossRef]
- Asiamah, E.K.; Vailati-Riboni, M.; Zhou, Z.; Xu, T.; Loor, J.J.; Schimmel, K.; Worku, M. Rumen-Protected Methionine Supplementation during the Peripartal Period Alters the Expression of Galectin Genes Associated with Inflammation in Peripheral Neutrophils and Secretion in Plasma of Holstein Cows. J. Dairy Res. 2019, 86, 394–398. [Google Scholar] [CrossRef]
- Osorio, J.S.; Jacometo, C.B.; Zhou, Z.; Luchini, D.; Cardoso, F.C.; Loor, J.J. Hepatic Global DNA and Peroxisome Proliferator-Activated Receptor Alpha Promoter Methylation Are Altered in Peripartal Dairy Cows Fed Rumen-Protected Methionine. J. Dairy Sci. 2016, 99, 234–244. [Google Scholar] [CrossRef]
- Zhou, Z.; Ferdous, F.; Montagner, P.; Luchini, D.N.; Corrêa, M.N.; Loor, J.J. Methionine and Choline Supply during the Peripartal Period Alter Polymorphonuclear Leukocyte Immune Response and Immunometabolic Gene Expression in Holstein Cows. J. Dairy Sci. 2018, 101, 10374–10382. [Google Scholar] [CrossRef]
- Hou, X.; Jiang, M.; Zhou, J.; Song, S.; Zhao, F.; Lin, Y. Examination of Methionine Stimulation of Gene Expression in Dairy Cow Mammary Epithelial Cells Using RNA-Sequencing. J. Dairy Res. 2020, 87, 226–231. [Google Scholar] [CrossRef]
- Duan, X.; Lin, Y.; Lv, H.; Yang, Y.; Jiao, H.; Hou, X. Methionine Induces LAT1 Expression in Dairy Cow Mammary Gland by Activating the MTORC1 Signaling Pathway. DNA Cell. Biol. 2017, 36, 1126–1133. [Google Scholar] [CrossRef]
- Yang, J.-X.; Wang, C.-H.; Xu, Q.-B.; Zhao, F.-Q.; Liu, J.-X.; Liu, H.-Y. Methionyl-Methionine Promotes α-S1 Casein Synthesis in Bovine Mammary Gland Explants by Enhancing Intracellular Substrate Availability and Activating JAK2-STAT5 and MTOR-Mediated Signaling Pathways. J. Nutr. 2015, 145, 1748–1753. [Google Scholar] [CrossRef]
- Qi, H.; Meng, C.; Jin, X.; Li, X.; Li, P.; Gao, X. Methionine Promotes Milk Protein and Fat Synthesis and Cell Proliferation via the SNAT2-PI3K Signaling Pathway in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2018, 66, 11027–11033. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. In Histone Mutations and Cancer; Advances in Experimental Medicine and Biology; Springer: Singapore, 2021; Volume 1283, pp. 1–16. [Google Scholar] [CrossRef]
- Sun, S.; Meng, Q.; Luo, Z.; Shi, B.; Bi, C.; Shan, A. Effects of Dietary Resveratrol Supplementation during Gestation and Lactation of Sows on Milk Composition of Sows and Fat Metabolism of Sucking Piglets. J. Anim. Physiol. Anim. Nutr. 2019, 103, 813–821. [Google Scholar] [CrossRef]
- Javaheri Barfourooshi, H.; Sadeghipanah, H.; Asadzadeh, N.; Seyedabadi, H.; Borazjani, M.; Javanmard, A. Changes in the Gene Expression Profile of the Mammary Gland Lipogenic Enzymes in Saanen Goats in Response to Dietary Fats. Vet. Med. Sci. 2023, 9, 945–956. [Google Scholar] [CrossRef]
- Sun, M.; Li, Z.; Xing, Y.; Mu, X.; Cao, Y.; Hao, Y.; Yang, J.; Li, D. Effects of Glucose Availability on AS1-Casein Synthesis in Bovine Mammary Epithelial Cells. J. Anim. Sci. 2022, 100, skac330. [Google Scholar] [CrossRef]
- Hosseini, A.; Tariq, M.R.; Trindade da Rosa, F.; Kesser, J.; Iqbal, Z.; Mora, O.; Sauerwein, H.; Drackley, J.K.; Trevisi, E.; Loor, J.J. Insulin Sensitivity in Adipose and Skeletal Muscle Tissue of Dairy Cows in Response to Dietary Energy Level and 2,4-Thiazolidinedione (TZD). PLoS ONE 2015, 10, e0142633. [Google Scholar] [CrossRef] [PubMed]
- Mann, S.; Leal Yepes, F.A.; Duplessis, M.; Wakshlag, J.J.; Overton, T.R.; Cummings, B.P.; Nydam, D.V. Dry Period Plane of Energy: Effects on Glucose Tolerance in Transition Dairy Cows. J. Dairy Sci. 2016, 99, 701–717. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Krause, D.O.; Gozho, G.N.; McBride, B.W. Subacute Ruminal Acidosis in Dairy Cows: The Physiological Causes, Incidence and Consequences. Vet. J. 2008, 176, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Nafikov, R.A.; Beitz, D.C. Carbohydrate and Lipid Metabolism in Farm Animals. J. Nutr. 2007, 137, 702–705. [Google Scholar] [CrossRef]
- Dong, G.; Qiu, M.; Ao, C.; Zhou, J.; Khas-Erdene; Wang, X.; Zhang, Z.; Yang, Y. Feeding a High-Concentrate Corn Straw Diet Induced Epigenetic Alterations in the Mammary Tissue of Dairy Cows. PLoS ONE 2014, 9, e107659. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ibeagha-Awemu, E.M. Altered Gene Expression of Epigenetic Modifying Enzymes in Response to Dietary Supplementation with Linseed Oil. J. Dairy Res. 2017, 84, 119–123. [Google Scholar] [CrossRef]
- Pomar, C.A.; Castillo, P.; Palou, A.; Palou, M.; Picó, C. Dietary Improvement during Lactation Normalizes MiR-26a, MiR-222 and MiR-484 Levels in the Mammary Gland, but Not in Milk, of Diet-Induced Obese Rats. Biomedicines 2022, 10, 1292. [Google Scholar] [CrossRef]
- Wicik, Z.; Gajewska, M.; Majewska, A.; Walkiewicz, D.; Osińska, E.; Motyl, T. Characterization of MicroRNA Profile in Mammary Tissue of Dairy and Beef Breed Heifers. J. Anim. Breed. Genet. 2016, 133, 31–42. [Google Scholar] [CrossRef]
- Wang, J.; Bian, Y.; Wang, Z.; Li, D.; Wang, C.; Li, Q.; Gao, X. MicroRNA-152 Regulates DNA Methyltransferase 1 and Is Involved in the Development and Lactation of Mammary Glands in Dairy Cows. PLoS ONE 2014, 9, e101358. [Google Scholar] [CrossRef]
- Bian, Y.; Lei, Y.; Wang, C.; Wang, J.; Wang, L.; Liu, L.; Liu, L.; Gao, X.; Li, Q. Epigenetic Regulation of MiR-29s Affects the Lactation Activity of Dairy Cow Mammary Epithelial Cells. J. Cell. Physiol. 2015, 230, 2152–2163. [Google Scholar] [CrossRef]
- Jiao, P.; Yuan, Y.; Zhang, M.; Sun, Y.; Wei, C.; Xie, X.; Zhang, Y.; Wang, S.; Chen, Z.; Wang, X. PRL/MicroRNA-183/IRS1 Pathway Regulates Milk Fat Metabolism in Cow Mammary Epithelial Cells. Genes 2020, 11, 196. [Google Scholar] [CrossRef]
- Roth, M.J.; Moorehead, R.A. The MiR-200 Family in Normal Mammary Gland Development. BMC Dev. Biol. 2021, 21, 12. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, S.; Zhang, Q.; Guo, X.; Wu, C.; Yao, M.; Sun, D. Comprehensive MicroRNA Expression Profile of the Mammary Gland in Lactating Dairy Cows With Extremely Different Milk Protein and Fat Percentages. Front. Genet. 2020, 11, 548268. [Google Scholar] [CrossRef]
- Billa, P.-A.; Faulconnier, Y.; Ye, T.; Bourdon, C.; Pires, J.A.A.; Leroux, C. Nutrigenomic Analyses Reveal MiRNAs and MRNAs Affected by Feed Restriction in the Mammary Gland of Midlactation Dairy Cows. PLoS ONE 2021, 16, e0248680. [Google Scholar] [CrossRef]
- Shore, A.N.; Kabotyanski, E.B.; Roarty, K.; Smith, M.A.; Zhang, Y.; Creighton, C.J.; Dinger, M.E.; Rosen, J.M. Pregnancy-Induced Noncoding RNA (PINC) Associates with Polycomb Repressive Complex 2 and Regulates Mammary Epithelial Differentiation. PLoS Genet. 2012, 8, e1002840. [Google Scholar] [CrossRef]
- Sun, P.; Chen, M.; Sooranna, S.R.; Shi, D.; Liu, Q.; Li, H. The Emerging Roles of CircRNAs in Traits Associated with Livestock Breeding. Wiley Interdiscip Rev. RNA 2023, e1775. [Google Scholar] [CrossRef]

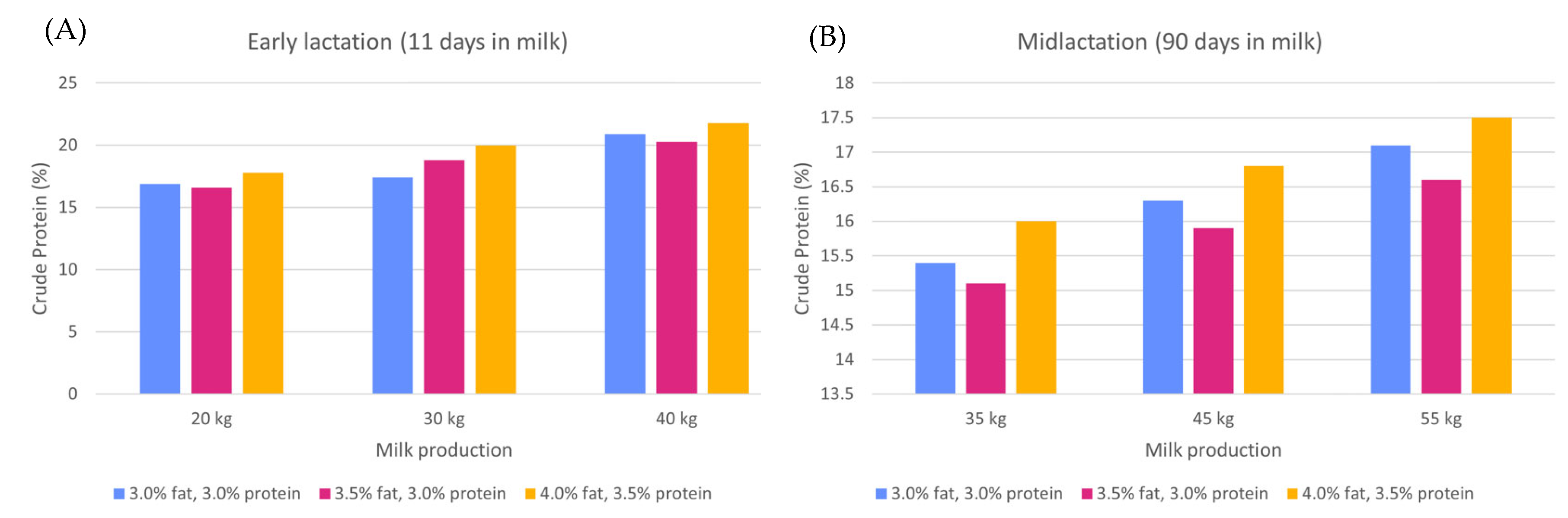
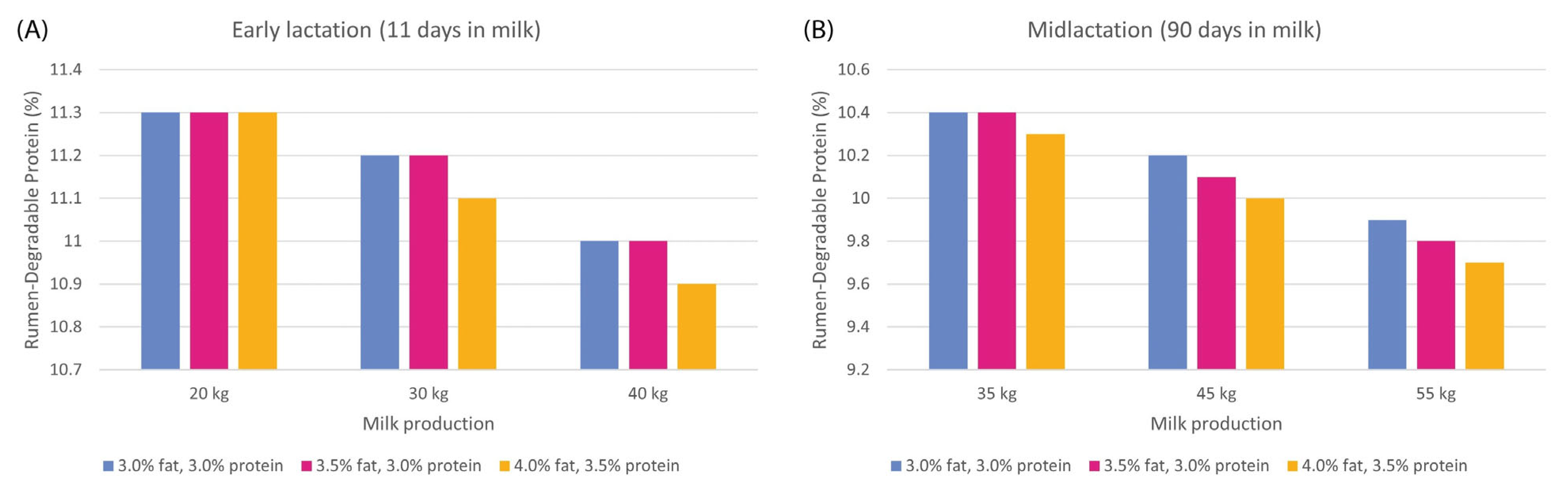

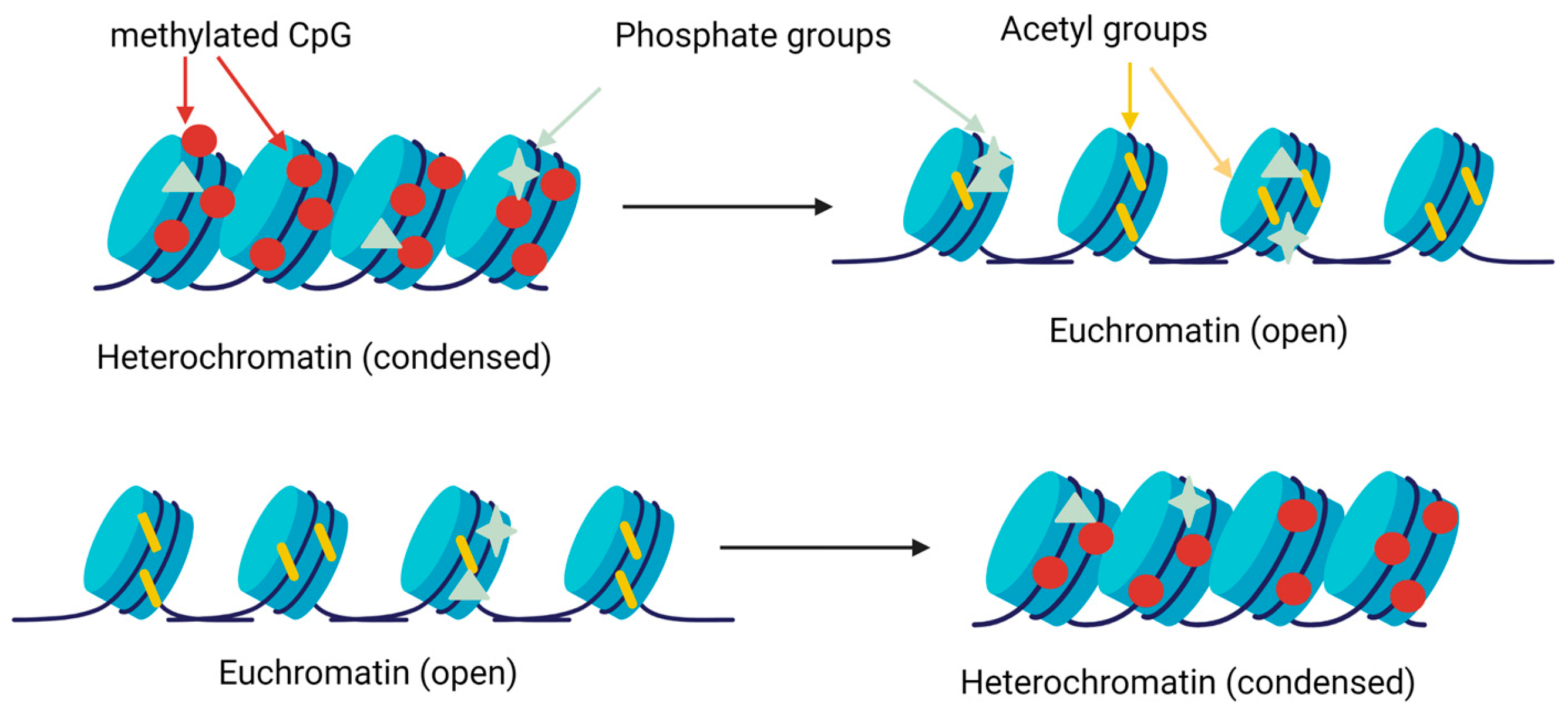
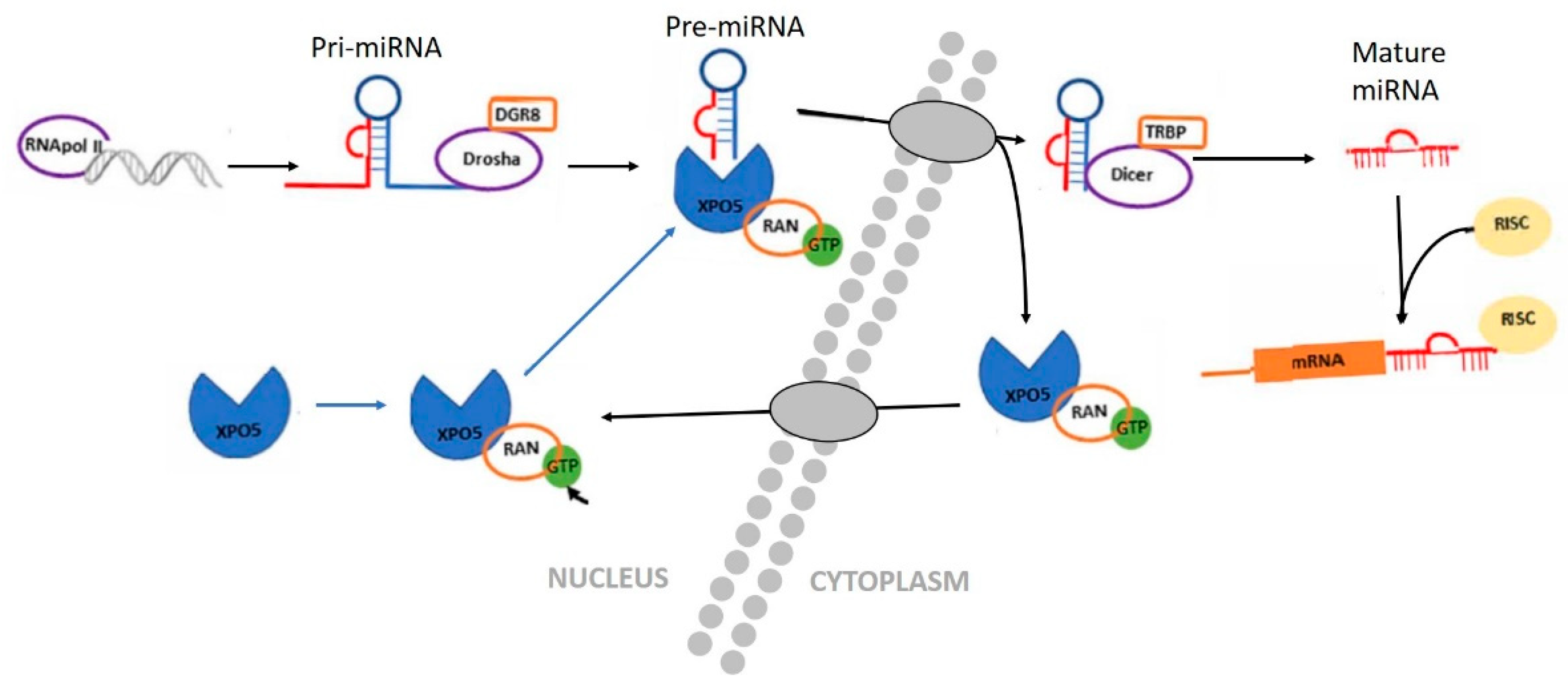
| Nutrient Examined | Epigenetic Mark or Molecule | Physiological Endpoint | References |
|---|---|---|---|
| Protein restriction | DNA methylation patterns altered | Milk production in cattle and goats | [85,86,87] |
| Methionine supplementation | Phosphorylation of NFE2L2 | Antioxidant effect in cattle | [90,91,92] |
| Upregulation of gene expression | Protein and fat synthesis in milk in cattle | [102,103,104] | |
| Choline supplementation | DNA methylation patterns altered | Milk production and liver fatty acid metabolism in cattle | [97] |
| Polyphenol and omega-3 fatty acid supplementation | Histone acetylation patterns | Milk composition in pigs and goats | [104,105] |
| Carbohydrates and fat supplementation | DNA methylation patterns altered | Reduction in milk fat content; milk production in cattle | [111,112] |
| High fat supplementation | MiRNA expression patterns altered | Milk production in rats | [115] |
| Energy restriction | MiRNA expression patterns altered | Milk production and composition in cattle | [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lesta, A.; Marín-García, P.J.; Llobat, L. How Does Nutrition Affect the Epigenetic Changes in Dairy Cows? Animals 2023, 13, 1883. https://doi.org/10.3390/ani13111883
Lesta A, Marín-García PJ, Llobat L. How Does Nutrition Affect the Epigenetic Changes in Dairy Cows? Animals. 2023; 13(11):1883. https://doi.org/10.3390/ani13111883
Chicago/Turabian StyleLesta, Ana, Pablo Jesús Marín-García, and Lola Llobat. 2023. "How Does Nutrition Affect the Epigenetic Changes in Dairy Cows?" Animals 13, no. 11: 1883. https://doi.org/10.3390/ani13111883
APA StyleLesta, A., Marín-García, P. J., & Llobat, L. (2023). How Does Nutrition Affect the Epigenetic Changes in Dairy Cows? Animals, 13(11), 1883. https://doi.org/10.3390/ani13111883








