Comparative Proteomic Analysis of Milk-Derived Extracellular Vesicles from Dairy Cows with Clinical and Subclinical Mastitis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Health and Milk Sampling
2.2. Preparation and Isolation of Milk-Derived EVs
2.3. Identification of Milk-Derived EVs
2.4. Protein Extraction and Digestion
2.5. LC-MS/MS Analysis
2.6. DIA Mode
2.7. Raw Data Processing
2.8. Bioinformatics Analysis
3. Results
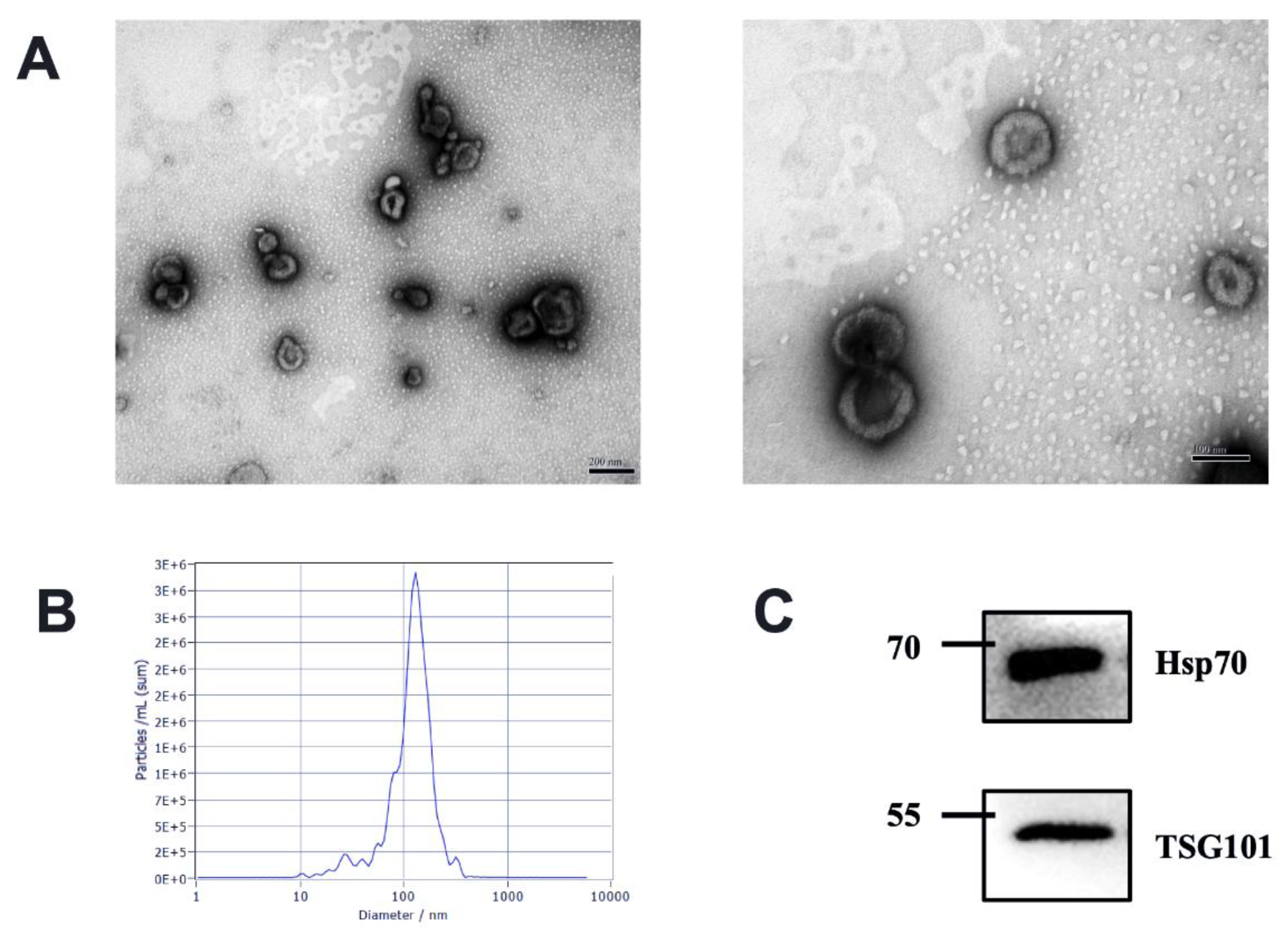
3.1. Characteristics of Milk-Derived EVs
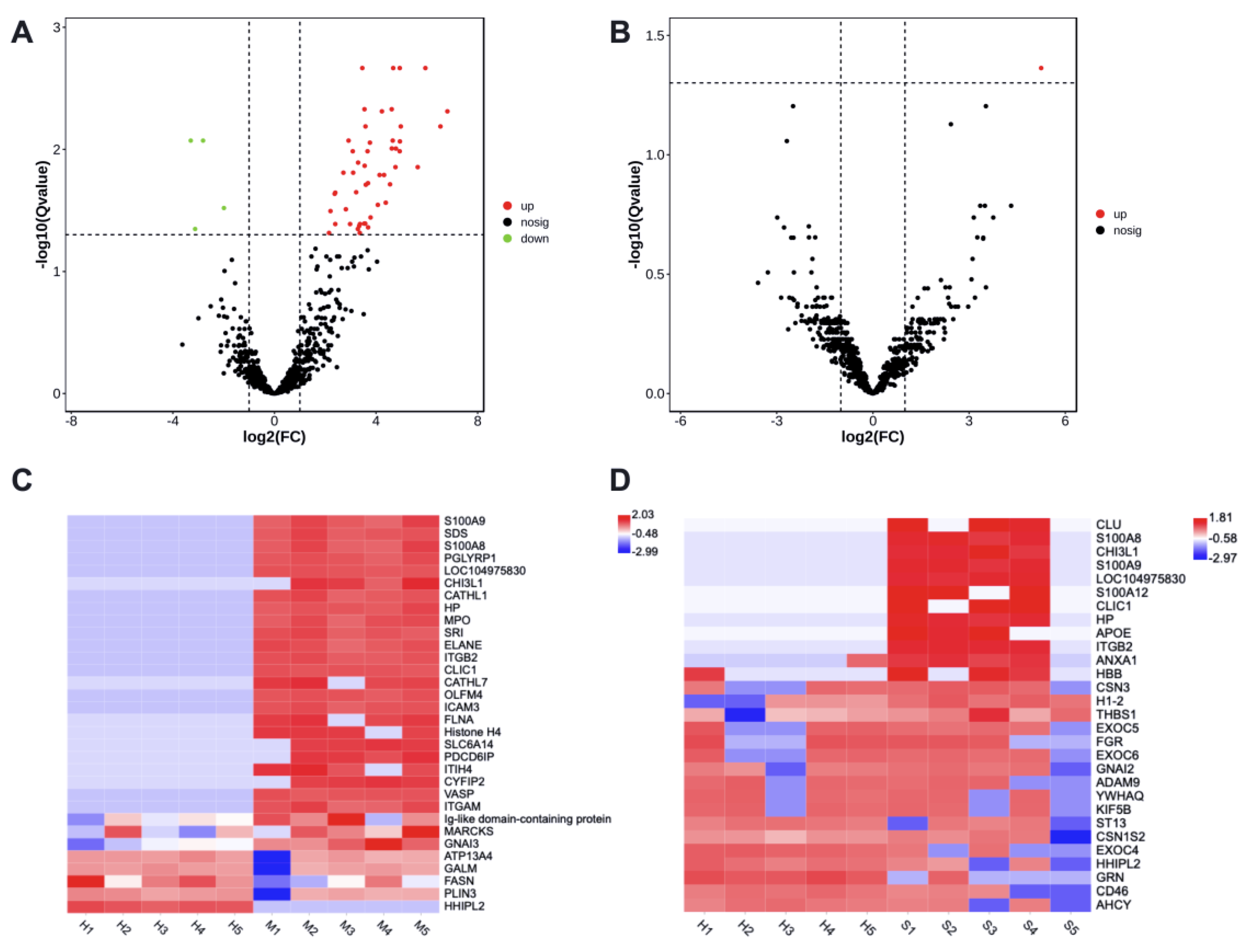
3.2. Statistical Analysis of Identified Proteins from Milk-Derived EVs
3.3. Mastitis-Associated Alterations in Proteome of Milk-Derived EVs
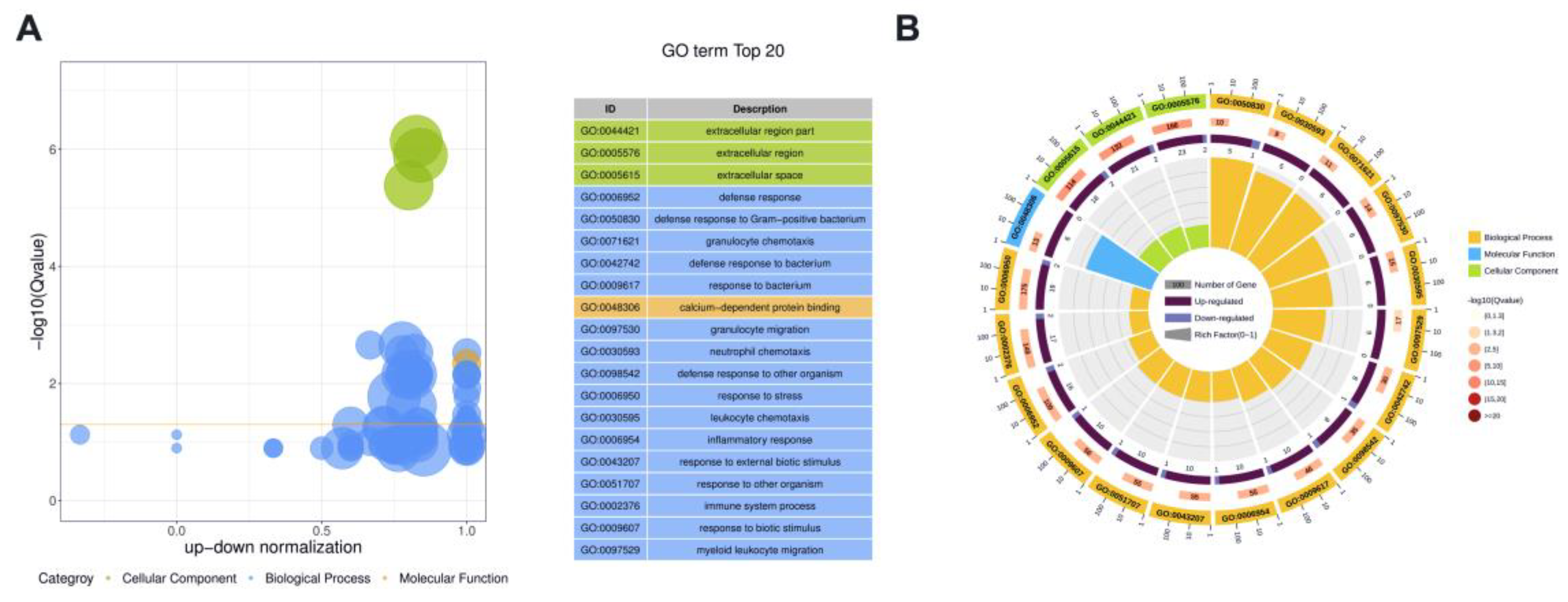
3.4. Functional Analysis of DE Proteins in Milk-Derived EVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human Saliva, Plasma and Breast Milk Exosomes Contain RNA: Uptake by Macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef]
- Foster, B.P.; Balassa, T.; Benen, T.D.; Dominovic, M.; Elmadjian, G.K.; Florova, V.; Fransolet, M.D.; Kestlerova, A.; Kmiecik, G.; Kostadinova, I.A.; et al. Extracellular Vesicles in Blood, Milk and Body Fluids of the Female and Male Urogenital Tract and with Special Regard to Reproduction. Crit. Rev. Clin. Lab. Sci. 2016, 53, 379–395. [Google Scholar] [CrossRef]
- Reinhardt, T.A.; Lippolis, J.D.; Nonnecke, B.J.; Sacco, R.E. Bovine Milk Exosome Proteome. J. Proteom. 2012, 75, 1486–1492. [Google Scholar] [CrossRef]
- Wolf, T.; Baier, S.R.; Zempleni, J. The Intestinal Transport of Bovine Milk Exosomes Is Mediated by Endocytosis in Human Colon Carcinoma Caco-2 Cells and Rat Small Intestinal IEC-6 Cells. J. Nutr. 2015, 145, 2201–2206. [Google Scholar] [CrossRef]
- Somiya, M.; Yoshioka, Y.; Ochiya, T. Biocompatibility of Highly Purified Bovine Milk-Derived Extracellular Vesicles. J. Extracell. Vesicles 2018, 7, 1440132. [Google Scholar] [CrossRef]
- Feng, X.; Chen, X.; Zheng, X.; Zhu, H.; Qi, Q.; Liu, S.; Zhang, H.; Che, J. Latest Trend of Milk Derived Exosomes: Cargos, Functions, and Applications. Front. Nutr. 2021, 8, 747294. [Google Scholar] [CrossRef]
- Zempleni, J.; Sukreet, S.; Zhou, F.; Wu, D.; Mutai, E. Milk-Derived Exosomes and Metabolic Regulation. Annu. Rev. Anim. Biosci. 2018, 7, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.; Atalla, H.; Karrow, N.; Mallard, B.A. The Bioactivity of Colostrum and Milk Exosomes of High, Average, and Low Immune Responder Cows on Human Intestinal Epithelial Cells. J. Dairy Sci. 2021, 104, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. A 100-Year Review: Mastitis Detection, Management, and Prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef]
- Martins, L.; Barcelos, M.M.; Cue, R.I.; Anderson, K.L.; Dos Santos, M.V.; Gonçalves, J.L. Chronic Subclinical Mastitis Reduces Milk and Components Yield at the Cow Level. J. Dairy Res. 2020, 87, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Busanello, M.; Rossi, R.S.; Cassoli, L.D.; Pantoja, J.C.F.; Machado, P.F. Estimation of Prevalence and Incidence of Subclinical Mastitis in a Large Population of Brazilian Dairy Herds. J. Dairy Sci. 2017, 100, 6545–6553. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.; Chisanga, D.; Liem, M.; Keerthikumar, S.; Anand, S.; Ang, C.-S.; Adda, C.G.; Versteegen, E.; Jois, M.; Mathivanan, S. Bovine Milk-Derived Exosomes from Colostrum Are Enriched with Proteins Implicated in Immune Response and Growth. Sci. Rep. 2017, 7, 5933. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Song, D.; Cao, X.; Wu, R.; Liu, B.; Ye, W.; Wu, J.; Yue, X. Comparative Proteomic Analysis of Milk-Derived Exosomes in Human and Bovine Colostrum and Mature Milk Samples by ITRAQ-Coupled LC-MS/MS. Food Res. Int. 2017, 92, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine Milk Proteome: Quantitative Changes in Normal Milk Exosomes, Milk Fat Globule Membranes and Whey Proteomes Resulting from Staphylococcus Aureus Mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef]
- Cai, M.; He, H.; Jia, X.; Chen, S.; Wang, J.; Shi, Y.; Liu, B.; Xiao, W.; Lai, S. Genome-Wide MicroRNA Profiling of Bovine Milk-Derived Exosomes Infected with Staphylococcus Aureus. Cell Stress Chaperones 2018, 23, 663–672. [Google Scholar] [CrossRef]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA Expression Profiles of Bovine Milk Exosomes in Response to Staphylococcus Aureus Infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef]
- Ma, S.; Tong, C.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and Characterization of Differentially Expressed Exosomal MicroRNAs in Bovine Milk Infected with Staphylococcus Aureus. BMC Genom. 2019, 20, 934. [Google Scholar] [CrossRef] [PubMed]
- Saenz-de-Juano, M.D.; Silvestrelli, G.; Bauersachs, S.; Ulbrich, S.E. Determining Extracellular Vesicles Properties and MiRNA Cargo Variability in Bovine Milk from Healthy Cows and Cows Undergoing Subclinical Mastitis. BMC Genom. 2022, 23, 189. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Chen, L.; Xiong, B.; Kang, B.; Zhang, P.; Tang, S.; Han, H.; Shen, W.; Feng, X.; Feng, S.; et al. Single-Cell Transcriptome Sequencing and Proteomics Reveal Neonatal Ileum Dynamic Developmental Potentials. Msystems 2021, 6, e00725-21. [Google Scholar] [CrossRef]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Braunstein, Z.; Toomey, A.C.; Zhong, J.; Rao, X. S100 Proteins As an Important Regulator of Macrophage Inflammation. Front. Immunol. 2018, 8, 1908. [Google Scholar] [CrossRef]
- Vogl, T.; Gharibyan, A.L.; Morozova-Roche, L.A. Pro-Inflammatory S100A8 and S100A9 Proteins: Self-Assembly into Multifunctional Native and Amyloid Complexes. Int. J. Mol. Sci. 2012, 13, 2893. [Google Scholar] [CrossRef]
- Achouiti, A.; Vogl, T.; Van der Meer, A.J.; Stroo, I.; Florquin, S.; de Boer, O.J.; Roth, J.; Zeerleder, S.; van′t Veer, C.; de Vos, A.F.; et al. Myeloid-Related Protein-14 Deficiency Promotes Inflammation in Staphylococcal Pneumonia. Eur. Respir. J. 2015, 46, 464–473. [Google Scholar] [CrossRef]
- Sprenkeler, E.G.; Zandstra, J.; van Kleef, N.D.; Goetschalckx, I.; Verstegen, B.; Aarts, C.E.; Janssen, H.; Tool, A.T.; van Mierlo, G.; van Bruggen, R.; et al. S100A8/A9 Is a Marker for the Release of Neutrophil Extracellular Traps and Induces Neutrophil Activation. Cells 2022, 11, 236. [Google Scholar] [CrossRef]
- Wartha, F.; Beiter, K.; Normark, S.; Henriques-Normark, B. Neutrophil Extracellular Traps: Casting the NET over Pathogenesis. Curr. Opin. Microbiol. 2007, 10, 52–56. [Google Scholar] [CrossRef]
- Prieto, D.; Sotelo, N.; Seija, N.; Sernbo, S.; Abreu, C.; Durán, R.; Gil, M.; Sicco, E.; Irigoin, V.; Oliver, C.; et al. S100-A9 Protein in Exosomes from Chronic Lymphocytic Leukemia Cells Promotes NF-ΚB Activity during Disease Progression. Blood 2017, 130, 777–788. [Google Scholar] [CrossRef]
- Li, H.; Huang, X.; Chang, X.; Yao, J.; He, Q.; Shen, Z.; Ji, Y.; Wang, K. S100-A9 Protein in Exosomes Derived from Follicular Fluid Promotes Inflammation via Activation of NF-κB Pathway in Polycystic Ovary Syndrome. J. Cell. Mol. Med. 2020, 24, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Lacy, P. Mechanisms of Degranulation in Neutrophils. Allergy Asthma Clin. Immunol. 2006, 2, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Huang, H.; Hu, M.; Wang, Q.; Gao, Y.; Liu, Y. Time-Dependent Expression of Leukotriene B4 Receptors in Rat Collagen-Induced Arthritis. Prostaglandins Other Lipid Mediat. 2007, 83, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.L.; Hall, A. Rho GTPases and Their Effector Proteins. Biochem. J. 2000, 348 Pt 2, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Günther, J.; Petzl, W.; Bauer, I.; Ponsuksili, S.; Zerbe, H.; Schuberth, H.-J.; Brunner, R.M.; Seyfert, H.-M. Differentiating Staphylococcus Aureus from Escherichia Coli Mastitis: S. Aureus Triggers Unbalanced Immune-Dampening and Host Cell Invasion Immediately after Udder Infection. Sci. Rep. 2017, 7, 4811. [Google Scholar] [CrossRef]
- Benedyk, M.; Sopalla, C.; Nacken, W.; Bode, G.; Melkonyan, H.; Banfi, B.; Kerkhoff, C. HaCaT Keratinocytes Overexpressing the S100 Proteins S100A8 and S100A9 Show Increased NADPH Oxidase and NF-ΚB Activities. J. Investig. Dermatol. 2007, 127, 2001–2011. [Google Scholar] [CrossRef]
- Nardo, A.D.; Braff, M.H.; Taylor, K.R.; Na, C.; Granstein, R.D.; McInturff, J.E.; Krutzik, S.; Modlin, R.L.; Gallo, R.L. Cathelicidin Antimicrobial Peptides Block Dendritic Cell TLR4 Activation and Allergic Contact Sensitization. J. Immunol. 2007, 178, 1829–1834. [Google Scholar] [CrossRef]
- Dziarski, R.; Platt, K.A.; Gelius, E.; Steiner, H.; Gupta, D. Defect in Neutrophil Killing and Increased Susceptibility to Infection with Nonpathogenic Gram-positive Bacteria in Peptidoglycan Recognition Protein-S (PGRP-S)–Deficient Mice. Blood 2003, 102, 689–697. [Google Scholar] [CrossRef]
- Rathnayake, N.; Gustafsson, A.; Sorsa, T.; Norhammar, A.; Bostanci, N.; PAROKRANK Steering Committee. Association of Peptidoglycan Recognition Protein 1 to Post-myocardial Infarction and Periodontal Inflammation: A Subgroup Report from the PAROKRANK (Periodontal Disease and the Relation to Myocardial Infarction) Study. J. Periodontol. 2022, 93, 1325–1335. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.-Y.; Roth, A.L.; Huang, B.Q.; Ward, H.D.; Zhu, G.; LaRusso, N.F.; Hanson, N.D.; Chen, X.-M. Release of Luminal Exosomes Contributes to TLR4-Mediated Epithelial Antimicrobial Defense. PLoS Pathog. 2013, 9, e1003261. [Google Scholar] [CrossRef]
- Wang, X.; Han, Y.; Liu, J.; Zhang, Y.; Cheng, K.; Guo, J.; Guo, Q.; Liu, S.; Sun, H.; Hua, Y.; et al. Exosomes Play an Important Role in the Progression of Plasma Cell Mastitis via the PI3K-Akt-MTOR Signaling Pathway. Mediat. Inflamm. 2019, 2019, 4312016. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.N.; Hu, H.; Wen, P.C.; Lian, S.; Xie, X.L.; Song, H.L.; Yang, Z.N.; Ren, F.Z. Yak Milk-Derived Exosomes Alleviate Lipopolysaccharide-Induced Intestinal Inflammation by Inhibiting PI3K/AKT/C3 Pathway Activation. J. Dairy Sci. 2021, 104, 8411–8424. [Google Scholar] [CrossRef] [PubMed]

| Uniport IDs | GeneNames | Protein Names | logFC | p Value | Adjust p |
|---|---|---|---|---|---|
| F1MHS5 | S100A9 | Protein S100-A9 | 6.81 | 0.000 | 0.005 |
| A0A3Q1LTZ6 | SDS | L-serine dehydratase/L-threonine deaminase | 6.54 | 0.000 | 0.006 |
| P28782 | S100A8 | Protein S100-A8 | 5.94 | 0.000 | 0.002 |
| Q8SPP7 | PGLYRP1 | Peptidoglycan recognition protein 1 | 4.94 | 0.000 | 0.009 |
| F1N1Z8 | LOC104975830 | Uncharacterized protein | 4.93 | 0.000 | 0.010 |
| A0A3Q1MT29 | CHI3L1 | Chitinase-3-like protein 1 | 4.76 | 0.000 | 0.014 |
| P22226 | CATHL1 | Cathelicidin-1 | 4.66 | 0.000 | 0.008 |
| Q2TBU0 | HP | Haptoglobin | 4.62 | 0.000 | 0.005 |
| A6QPT4 | MPO | MPO protein | 4.23 | 0.000 | 0.005 |
| Q0IIA3 | SRI | Sorcin | 3.76 | 0.000 | 0.009 |
| A6QPP7 | ELANE | ELA2 protein | 3.71 | 0.008 | 0.096 |
| P32592 | ITGB2 | Integrin beta-2 | 3.54 | 0.000 | 0.005 |
| Q5E9B7 | CLIC1 | Chloride intracellular channel protein 1 | 3.46 | 0.000 | 0.002 |
| P56425 | CATHL7 | Cathelicidin-7 | 3.37 | 0.002 | 0.041 |
| F1MVJ8 | OLFM4 | Olfactomedin 4 | 3.29 | 0.000 | 0.013 |
| F1MQF0 | ICAM3 | Intercellular adhesion molecule 3 | 3.22 | 0.001 | 0.022 |
| F1N169 | FLNA | Filamin A | 3.15 | 0.005 | 0.077 |
| A0A3Q1M1Z1 | Histone H4 | Histone H4 | 3.10 | 0.001 | 0.016 |
| A5D7H9 | SLC6A14 | Transporter | 2.88 | 0.008 | 0.094 |
| E1BKM4 | PDCD6IP | Programmed cell death 6 interacting protein | 2.71 | 0.000 | 0.016 |
| A0A3Q1MA31 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 | 2.55 | 0.005 | 0.075 |
| F1MX60 | CYFIP2 | Cytoplasmic FMR1-interacting protein | 2.39 | 0.001 | 0.023 |
| Q2TA49 | VASP | Vasodilator-stimulated phosphoprotein | 1.85 | 0.014 | 0.148 |
| A0A3Q1M1Z4 | Ig-like | Ig-like domain-containing protein | 1.69 | 0.046 | 0.296 |
| P12624 | MARCKS | Myristoylated alanine-rich C-kinase substrate | 1.66 | 0.008 | 0.096 |
| G3MYD9 | ITGAM | Integrin subunit alpha M | 1.64 | 0.032 | 0.241 |
| Q3ZCA7 | GNAI3 | G protein subunit alpha i3 | 1.36 | 0.020 | 0.186 |
| F1N272 | ATP13A4 | Cation-transporting ATPase | −1.11 | 0.036 | 0.256 |
| Q5EA79 | GALM | Aldose 1-epimerase | −1.18 | 0.047 | 0.296 |
| F1N647 | FASN | Fatty acid synthase | −1.32 | 0.030 | 0.237 |
| Q3SX32 | PLIN3 | Perilipin | −1.99 | 0.001 | 0.030 |
| E1BGX8 | HHIPL2 | HHIP like 2 | −2.81 | 0.000 | 0.008 |
| Uniport IDs | Gene Names | Protein Names | logFC | p Value | Adjust p |
|---|---|---|---|---|---|
| P17697 | CLU | Clusterin | 3.75 | 0.002 | 0.183 |
| P28782 | S100A8 | Protein S100-A8 | 3.49 | 0.002 | 0.163 |
| A0A3Q1MT29 | CHI3L1 | Chitinase-3-like protein 1 | 3.44 | 0.006 | 0.225 |
| F1MHS5 | S100A9 | Protein S100-A9 | 3.18 | 0.021 | 0.396 |
| F1N1Z8 | LOC104975830 | Uncharacterized protein | 3.07 | 0.011 | 0.332 |
| P79105 | S100A12 | Protein S100-A12 | 2.97 | 0.034 | 0.432 |
| Q5E9B7 | CLIC1 | Chloride intracellular channel protein 1 | 2.43 | 0.000 | 0.074 |
| Q2TBU0 | HP | Haptoglobin | 2.26 | 0.014 | 0.359 |
| A0A3Q1LPF0 | APOE | Apolipoprotein E | 1.93 | 0.049 | 0.488 |
| P32592 | ITGB2 | Integrin beta-2 | 1.71 | 0.015 | 0.363 |
| F1N650 | ANXA1 | Annexin | 2.19 | 0.046 | 0.488 |
| P02070 | HBB | Hemoglobin subunit beta | 2.51 | 0.045 | 0.488 |
| A0A3Q1M5U9 | CSN3 | Kappa-casein | 2.57 | 0.034 | 0.432 |
| P02253 | H1-2 | Histone H1.2 | 3.11 | 0.007 | 0.273 |
| F1N3A1 | THBS1 | Thrombospondin-1 | 3.44 | 0.005 | 0.222 |
| F1MC71 | EXOC5 | Exocyst complex component 5 | −1.28 | 0.020 | 0.396 |
| A5PKG9 | FGR | Tyrosine-protein kinase | −1.76 | 0.040 | 0.471 |
| A0A3Q1M4P7 | EXOC6 | Exocyst complex component | −2.22 | 0.039 | 0.471 |
| A7MBH9 | GNAI2 | G protein subunit alpha i2 | 1.61 | 0.015 | 0.363 |
| F1MZJ5 | ADAM9 | ADAM metallopeptidase domain 9 | −1.03 | 0.041 | 0.474 |
| Q3SZI4 | YWHAQ | 14-3-3 protein theta | −1.79 | 0.043 | 0.482 |
| F1N1G7 | KIF5B | Kinesin-like protein | −1.82 | 0.049 | 0.488 |
| A7E3S8 | ST13 | Heat shock 70kD protein binding protein | −1.45 | 0.028 | 0.429 |
| P02663 | CSN1S2 | Alpha-S2-casein | 1.40 | 0.022 | 0.397 |
| A6QLD1 | EXOC4 | EXOC4 protein | −1.80 | 0.004 | 0.222 |
| E1BGX8 | HHIPL2 | HHIP like 2 | −2.00 | 0.003 | 0.199 |
| E1BHY6 | GRN | Granulin precursor | −2.07 | 0.032 | 0.432 |
| A0A3Q1LW07 | CD46 | Membrane cofactor protein | −2.49 | 0.000 | 0.063 |
| Q3MHL4 | AHCY | Adenosylhomocysteinase | −3.28 | 0.009 | 0.311 |
| Pathway Name | Entities | Reactions | ||||||
|---|---|---|---|---|---|---|---|---|
| Found | Total | Ratio | p Value | FDR | Found | Total | Ratio | |
| Neutrophil degranulation | 16 | 486 | 0.056 | 0.00000002 | 0.000004 | 9 | 10 | 0.001 |
| Innate Immune System | 20 | 901 | 0.104 | 0.00000014 | 0.000015 | 54 | 429 | 0.054 |
| Antimicrobial peptides | 5 | 56 | 0.006 | 0.00002490 | 0.001790 | 6 | 38 | 0.005 |
| Immune System | 21 | 1584 | 0.183 | 0.00022800 | 0.012300 | 56 | 1037 | 0.131 |
| RHO GTPases Activate NADPH Oxidases | 3 | 22 | 0.003 | 0.00034800 | 0.015000 | 9 | 14 | 0.002 |
| Metal sequestration by antimicrobial proteins | 2 | 7 | 0.001 | 0.00087400 | 0.029000 | 2 | 5 | 0.001 |
| RHO GTPases Activate WASPs and WAVEs | 3 | 31 | 0.004 | 0.00093600 | 0.029000 | 6 | 7 | 0.001 |
| RHO GTPase Effectors | 7 | 287 | 0.033 | 0.00176000 | 0.047600 | 22 | 88 | 0.011 |
| Regulation of actin dynamics for phagocytic cup formation | 3 | 53 | 0.006 | 0.00426000 | 0.102000 | 7 | 18 | 0.002 |
| Regulation of TLR by endogenous ligand | 2 | 17 | 0.002 | 0.00496000 | 0.104000 | 1 | 11 | 0.001 |
| EPH-Ephrin signaling | 3 | 65 | 0.007 | 0.00746000 | 0.142000 | 2 | 32 | 0.004 |
| Fcgamma receptor (FCGR) dependent phagocytosis | 3 | 67 | 0.008 | 0.00810000 | 0.146000 | 7 | 29 | 0.004 |
| Toll-like Receptor Cascades | 4 | 140 | 0.016 | 0.01060000 | 0.165000 | 3 | 116 | 0.015 |
| Neurofascin interactions | 1 | 2 | 0 | 0.01220000 | 0.165000 | 1 | 1 | 0 |
| Events associated with phagocytolytic activity of PMN cells | 1 | 2 | 0 | 0.01220000 | 0.165000 | 4 | 5 | 0.001 |
| RIPK1-mediated regulated necrosis | 2 | 29 | 0.003 | 0.01380000 | 0.165000 | 2 | 22 | 0.003 |
| Regulation of necroptotic cell death | 2 | 29 | 0.003 | 0.01380000 | 0.165000 | 1 | 16 | 0.002 |
| EPHB-mediated forward signaling | 2 | 31 | 0.004 | 0.01560000 | 0.184000 | 1 | 11 | 0.001 |
| Signaling by Rho GTPases, Miro GTPases and RHOBTB3 | 9 | 653 | 0.075 | 0.01670000 | 0.184000 | 30 | 176 | 0.022 |
| ROS and RNS production in phagocytes | 2 | 37 | 0.004 | 0.02170000 | 0.217000 | 5 | 10 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Cai, M.; Zhu, X.; Nan, X.; Xiong, B.; Yang, L. Comparative Proteomic Analysis of Milk-Derived Extracellular Vesicles from Dairy Cows with Clinical and Subclinical Mastitis. Animals 2023, 13, 171. https://doi.org/10.3390/ani13010171
Wang M, Cai M, Zhu X, Nan X, Xiong B, Yang L. Comparative Proteomic Analysis of Milk-Derived Extracellular Vesicles from Dairy Cows with Clinical and Subclinical Mastitis. Animals. 2023; 13(1):171. https://doi.org/10.3390/ani13010171
Chicago/Turabian StyleWang, Mengling, Meng Cai, Xiaoyan Zhu, Xuemei Nan, Benhai Xiong, and Liang Yang. 2023. "Comparative Proteomic Analysis of Milk-Derived Extracellular Vesicles from Dairy Cows with Clinical and Subclinical Mastitis" Animals 13, no. 1: 171. https://doi.org/10.3390/ani13010171
APA StyleWang, M., Cai, M., Zhu, X., Nan, X., Xiong, B., & Yang, L. (2023). Comparative Proteomic Analysis of Milk-Derived Extracellular Vesicles from Dairy Cows with Clinical and Subclinical Mastitis. Animals, 13(1), 171. https://doi.org/10.3390/ani13010171





