Activities of Antioxidant and Proteolytic Systems and Biomarkers in the Fat Body and Hemolymph of Young Apis mellifera Females
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Hemolymph and Fat Body Collection
2.3. Biochemical Analyses
2.3.1. Determination of Proteolytic System Activities
- Proteolytic activity test in relation to substrates (gelatine, haemoglobin, ovalbumin, albumin, cytochrome C, casein): 1 µL of each supernatant was incubated with 2 µL of each of the six substrates (1%, w/v) in an appropriate buffer for 120 min at 37 °C. The reactions were ended by adding 8 µL of cold 5% trichloroacetic acid (TCA). The supernatant was analysed spectrophotometrically (Synergy HTX (S1LFA); Warsaw, Poland) to measure the absorbance at 280 nm. The results obtained in this way allowed us to choose haemoglobin as the optimal substrate for further analyses. For more methodological details, see the Anson method [17] modified by Strachecka et al. [18,19].
- The activities of acidic, neutral, and alkaline proteases were assayed in three buffers, respectively: 100 mM glycine–HCl at pH 2.4, 100 mM Tris–HCl at pH 7.0, and 100 mM glycine–NaOH at pH 11.2. Next, 1 µL of each supernatant was incubated with 2 µL of 1% (w/v) haemoglobin in an appropriate buffer for 90 min at 37 °C. The reactions were ended by adding 8 µL of cold 5% trichloroacetic acid (TCA); the undigested proteins were precipitated and centrifuged for 1 min at 17,709× g rcf. The supernatant was spectrophotometrically analysed to measure the absorbance at 280 nm. One unit of enzyme activity was defined as the number of enzymes producing a 0.001 increase in absorbance per minute, according to Anson [17]. For more methodological details, see the Anson method [17] modified by Strachecka et al. [18,19].
- Determination of the activities of natural inhibitors of acidic, neutral, and alkaline proteases, based on the Lee and Lin method [20]. Pepsin was used as a marker for acidic, whereas trypsin was used for neutral and alkaline proteases. Then, 1 µL of pepsin or trypsin (1 mg/mL) was preincubated with 1 µL of a given supernatant for 30 min at 37 °C. After this time, 5 µL of 1% haemoglobin in an appropriate buffer were added, and the incubation was continued for 60 min. The reactions were terminated by adding 12 µL of trichloroacetic acid (TCA), centrifuged for 1 min at 17,709× g rcf and the supernatants were spectrophotometrically analysed to measure the absorbance at 280 nm. Inhibitor activities were calculated according to Lee and Lin [20].
- Proteolytic activities after the addition of pepstatin A, PMSF, iodoacetamide, and o-phenanthroline (the diagnostic inhibitors): 1 µL of diagnostic inhibitors (2 mM) was preincubated with 1 µL of a given supernatant for 30 min at 37 °C. After this time, 5 µL of 1% haemoglobin in an appropriate buffer were added, and the incubation continued for 90 min. The reactions were ended by adding 12 µL of trichloroacetic acid (TCA), and the supernatants were measured as described above. Proteolytic activities after the addition of the diagnostic inhibitors were calculated according to the Lee and Lin method [20].
2.3.2. Determination of Antioxidant System Activities
- Superoxide dismutase (SOD) determined using a commercial Sigma-Aldrich (19,160) SOD Determination Kit (Poznań, Poland);
- Catalase (CAT) determined using a Catalase Assay Kit (219265-1KIT) from Sigma-Aldrich (Poznań, Poland);
- Total antioxidant capacity (TAC) determined using a Total Antioxidant Capacity Assay Kit (MAK187-1KT) from Sigma-Aldrich (Poznań, Poland).
- All antioxidant enzyme activities were calculated per 1 mg of protein.
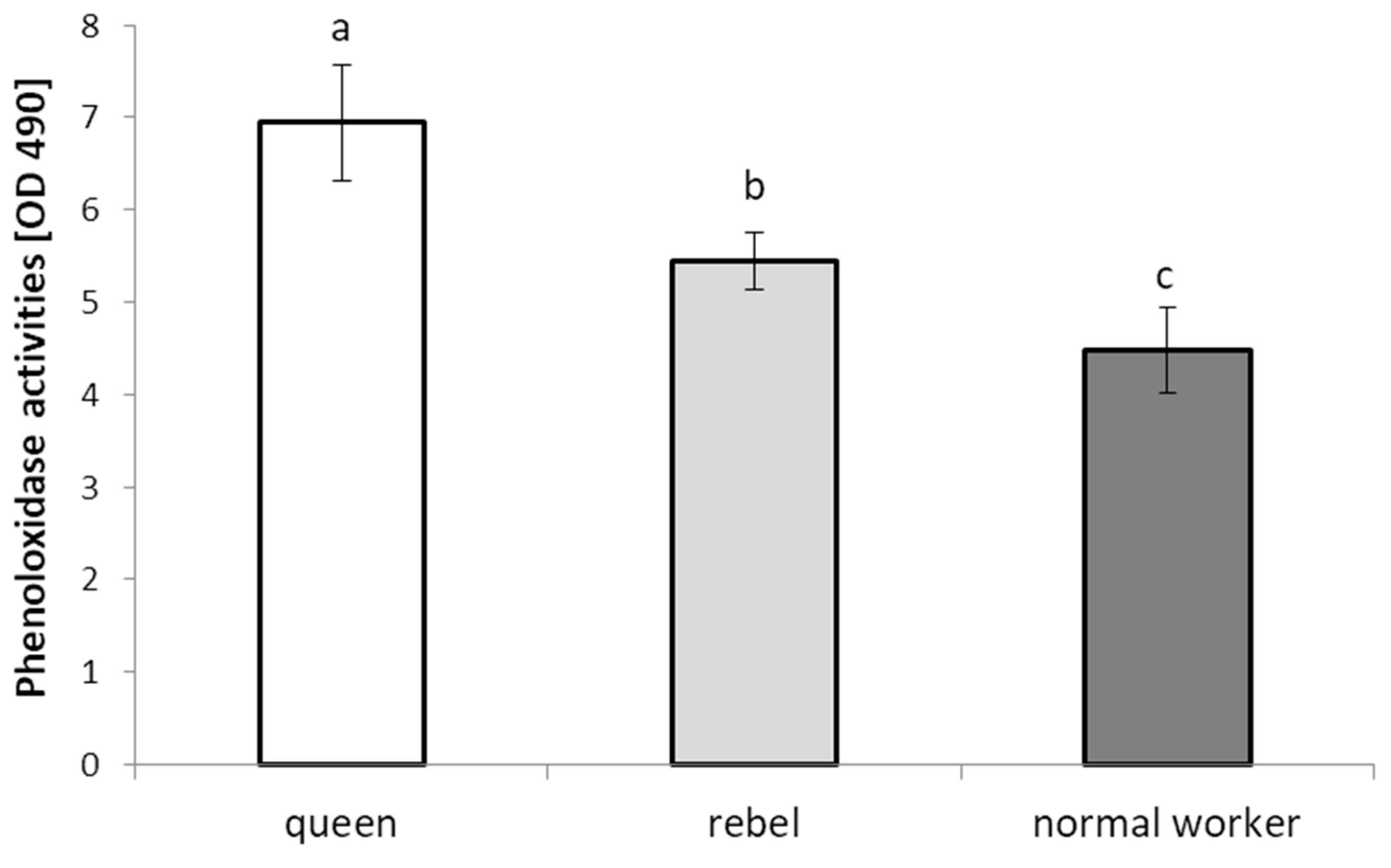
- The phenoloxidase (PO) activities were determined according to the method used by Ptaszyńska et al. [21]. Two microliters of the hemolymph solution were added to 18 μL of TBS (Cayman Chemical, Ann Arbor, MI, USA), containing 5 mM CaCl2 in the wells of a 96-well plate. After 20 min of incubation at room temperature, 180 μL of 2 mM L-dihydroxyphenylalanine (L-DOPA) in 50 mM sodium phosphate, pH 6.5, was added. PO activity was determined spectrophotometrically on the basis of the amount of melanin formed (absorbance at 490 nm) over 60 min, at 2-min intervals, using the Synergy HTX (BioTek, Janki, Poland) microplate reader. The PO activities were determined in triplicate for each hemolymph solution.
2.3.3. Determination of Biomarker Activities
2.4. Examination of Anatomical Characteristics
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borba, R.S.; Hoover, S.E.; Currie, R.W.; Giovenazzo, P.; Guarna, M.M.; Foster, L.J.; Zayed, A.; Permal, F.S. Phenomic analysis of the honey bee pathogen-web and its dynamics on colony productivity, health and social immunity behaviors. PLoS ONE 2022, 17, e0263273. [Google Scholar] [CrossRef] [PubMed]
- Papa, G.; Maier, R.; Durazzo, A.; Lucarini, M.; Karabagias, I.K.; Plutino, M.; Bianchetto, E.; Aromolo, R.; Pignatti, G.; Ambrogio, A.; et al. The honey bee Apis mellifera: An insect at the interface between human and ecosystem health. Biology 2022, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- Gallai, N.; Salles, J.M.; Settele, J.; Vaissière, B.E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 2009, 68, 810–821. [Google Scholar] [CrossRef]
- Grant, K.J.; DeVetter, L.; Melathopoulos, A. Honey bee (Apis mellifera) colony strength and its effects on pollination and yield in highbush blueberries (Vaccinium corymbosum). PeerJ 2021, 9, e11634. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.A.; Al-Ghamdi, M.S.; Ahmed, A.M.; Mohamed, A.S.A.; Shaker, G.H.; Ansari, M.J.; Dorrah, M.A.; Khan, K.A.; Ayaad, T.H. Immune investigation of the honeybee Apis mellifera jemenitica broods: A step toward production of a bee-derived antibiotic against the American foulbrood. Saudi J. Biol. Sci. 2021, 28, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Strachecka, A.; Migdał, P.; Kuszewska, K.; Skowronek, P.; Grabowski, M.; Paleolog, J.; Woyciechowski, M. Humoral and cellular defense mechanisms in rebel workers of Apis mellifera. Biology 2021, 10, 1146. [Google Scholar] [CrossRef]
- Grzywnowicz, K.; Ciołek, A.; Tabor, A.; Jaszek, M. Profiles of the body-surface proteolytic system of honey bee queens, workers and drones: Ontogenetic and seasonal changes in proteases and their natural inhibitors. Apidologie 2009, 40, 4–19. [Google Scholar] [CrossRef]
- Paleolog, J.; Wilde, J.; Miszczak, A.; Gancarz, M.; Strachecka, A. Antioxidation defenses of Apis mellifera queens and workers respond to imidacloprid in different age-dependent ways: Old queens are resistant, foragers are not. Animals 2021, 11, 1246. [Google Scholar] [CrossRef]
- Łoś, A.; Strachecka, A. Fast and cost-effective biochemical spectrophotometric analysis of solution of insect “blood” and body surface elution. Sensors 2018, 18, 1494. [Google Scholar] [CrossRef] [PubMed]
- Migdał, P.; Murawska, A.; Bieńkowski, P.; Strachecka, A.; Roman, A. Effect of the electric field at 50 Hz and variable intensities on biochemical markers in the honey bee’s hemolymph. PLoS ONE 2021, 16, e0252858. [Google Scholar] [CrossRef]
- Watson, F.; Püttmann-Holgado, R.; Thomas, F.; Lamar, D.; Hughes, M.; Kondo, M.; Rebel, V.I.; Schmucker, D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 2005, 309, 1874–1878. [Google Scholar] [CrossRef] [PubMed]
- Keehnen, N.L.P.; Fors, L.; Järver, P.; Spetz, A.; Nylin, S.; Theopold, U.; Wheat, C. A population genomic investigation of immune cell diversity and phagocytic capacity in a butterfly. Genes 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Strachecka, A.; Olszewski, K.; Kuszewska, K.; Chobotow, J.; Wójcik, Ł.; Paleolog, J.; Woyciechowski, M. Segmentation of the subcuticular fat body in Apis mellifera females with different reproductive potentials. Sci. Rep. 2021, 11, 13887. [Google Scholar] [CrossRef] [PubMed]
- Paes de Oliveira, V.T.; Poiani, S.B.; Antonialli, W.F.; Da Cruz-Landim, C. Morphometric changes on honeybee Apis mellifera L. workers fat body cells after juvenile hormone topic application at emergence. Micron 2008, 39, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Woyciechowski, M.; Kuszewska, K. Swarming generates rebel workers in honeybees. Curr. Biol. 2012, 22, 707–711. [Google Scholar] [CrossRef][Green Version]
- Büchler, R.; Andonov, S.; Bienefeld, K.; Costa, C.; Hatjina, F.; Kezic, N.; Kryger, P.; Spivak, M.; Uzunov, A.; Wilde, J. Standard methods for rearing and selection of Apis mellifera queens. J. Apic. Res. 2013, 52, 1–29. [Google Scholar] [CrossRef]
- Anson, M. The estimation of pepsin, trypsin, papain and cathepsin with hemoglobin. J. Gen. Physiol. 1938, 22, 79–84. [Google Scholar] [CrossRef]
- Strachecka, A.; Gryzińska, M.; Krauze, M. The influence of environmental pollution on the protective proteolytic barrier of the honey bee Apis mellifera mellifera. Pol. J. Environ. Stud. 2010, 19, 855–859. [Google Scholar]
- Strachecka, A.; Gryzińska, M.; Krauze, M.; Grzywnowicz, K. Profile of the body surface proteolytic system in Apis mellifera queens. Czech J. Anim. Sci. 2011, 56, 15–22. [Google Scholar] [CrossRef]
- Lee, T.; Lin, Y. Trypsin inhibitor and trypsin- like protease activity in air- or submergence-grown rice (Oryza sativa L.) coleoptiles. Plant Sci. 1995, 106, 43–54. [Google Scholar] [CrossRef]
- Ptaszyńska, A.A.; Borsuk, G.; Zdybicka-Barabas, A.; Cytryńska, M.; Małek, W. Are commercial probiotics and prebiotics effective in the treatment and prevention of honeybee nosemosis C? Parasitol. Res. 2016, 115, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Fang, Y.; Han, B.; Zhang, L.; Lu, X.; Li, J. Novel aspects of understanding molecular working mechanisms of salivary glands of worker honeybees (Apis mellifera) investigated by proteomics and phosphoproteomics. J. Proteom. 2013, 87, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bajda, M. Morphological Image and Biochemical Indicators in the Selected Tissues of Honey Bee Queens Depending on Their Age and Reproductive Status. Ph.D. Thesis, University of Life Sciences in Lublin, Lublin, Poland, 2017. [Google Scholar]
- Matsuoka, T.; Kawashima, T.; Nakamura, T.; Kanamaru, Y.; Yabe, T. Isolation and characterization of proteases that hydrolyze royal jelly proteins from queen bee larvae of the honeybee, Apis mellifera. Apidologie 2012, 43, 685–697. [Google Scholar] [CrossRef]
- Brandt, A.; Grikscheit, K.; Siede, R.; Grosse, R.; Meixner, M.D.; Büchler, R. Immunosuppression in honeybee queens by the neonicotinoids thiacloprid and clothianidin. Sci. Rep. 2017, 7, 4673. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.R.; Brockmann, A.; Pirk, C.W.W.; Stanley, D.W.; Tautz, J. Adult honeybees (Apis mellifera L.) abandon hemocytic, but not phenoloxidase-based immunity. J. Insect Physiol. 2008, 54, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.G.; Paxton, R.J.; Hernández-Sotomayor, S.M.T.; Pech-Jiménez, C.; Medina-Medina, L.A.; Quezada-Euán, J.J.G. Heat stress during development affects immunocompetence in workers, queens and drones of Africanized honey bees (Apis mellifera L.) (Hymenoptera: Apidae). J. Therm. Biol. 2020, 89, 102541. [Google Scholar] [CrossRef]
- Sagona, S.; Fronte, B.; Coppola, F.; Tafi, E.; Giusti, M.; Palego, L.; Betti, L.; Giannaccini, G.; Guglielminetti, L.; Felicioli, A. Effect of honey and syrup diets enriched with 1,3-1,6 β-glucans on honeybee survival rate and phenoloxidase activity (Apis mellifera L. 1758). Vet. Sci. 2021, 8, 130. [Google Scholar] [CrossRef]
- Ahmed, S.; Al Baki, M.; Lee, J.; Seo, D.; Lee, D.; Kim, Y. The first report of prostacyclin and its physiological roles in insects. Gen. Comp. Endocrinol. 2021, 301, 113659. [Google Scholar] [CrossRef]
- Koch, S.I.; Groh, K.; Vogel, H.; Hannson, B.S.; Kleineidam, C.J.; Grosse-Wilde, E. Caste-specific expression patterns of immune response and chemosensory related genes in the leaf-cutting ant, Atta vollenweideri. PLoS ONE 2013, 8, e81518. [Google Scholar] [CrossRef]
- Gao, L.; Wang, L.; Yang, X.; Wang, Y.; Liu, Z.; Xu, B.; Guo, X. Role of a serine protease gene (AccSp1) from Apis cerana cerana in abiotic stress responses and innate immunity. Cell Stress Chaperones 2019, 24, 29–43. [Google Scholar] [CrossRef]
- Strachecka, A.; Olszewski, K.; Paleolog, J. Varroa treatment with bromfenvinphos markedly suppresses honeybee biochemical defence levels. Entomol. Exp. Appl. 2016, 160, 57–71. [Google Scholar] [CrossRef]
- McAfee, A.; Chapman, A.; Pettis, J.S.; Foster, L.J.; Tarpy, D. Trade-offs between sperm viability and immune protein expression in honey bee queens (Apis mellifera). Commun. Biol. 2021, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Migdał, P.; Murawska, A.; Strachecka, A.; Bieńkowski, P.; Roman, A. Honey bee proteolytic system and behavior parameters under the influence of an electric field at 50 Hz and variable intensities for a long exposure time. Animals 2021, 11, 863. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W.; Chen, C.-H.; Hsu, C.-Y. Middle-aged worker bees express higher innate immunity than young worker bees in the abdomen without the digestive tract of worker bees reared in an incubator. Insects 2022, 13, 209. [Google Scholar] [CrossRef]
- Gábor, E.; Cinege, G.; Csordás, G.; Rusvai, M.; Honti, V.; Kolics, B.; Török, T.; Williams, M.J.; Kurucz, É.; Andó, I. Identification of reference markers for characterizing honey bee (Apis mellifera) hemocyte classes. Dev. Comp. Immunol. 2020, 109, 103701. [Google Scholar] [CrossRef]
- Costantini, D. Understanding diversity in oxidative status and oxidative stress: The opportunities and challenges ahead. J. Exp. Biol. 2019, 222, 13. [Google Scholar] [CrossRef]
- Linksvayer, T.A.; Wade, M.J. The evolutionary origin and elaboration of sociality in the aculeate Hymenoptera: Maternal effects, sib-social effects, and heterochrony. Q. Rev. Biol. 2005, 80, 317–336. [Google Scholar] [CrossRef]
- Sasaki, K.; Watanabe, T. Sex-specific regulatory systems for dopamine production in the honey bee. Insects 2022, 13, 128. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Hsieh, Y.-S. Oxidative stress decreases in the trophocytes and fat cells of worker honeybees during aging. Biogerontology 2013, 15, 129–137. [Google Scholar] [CrossRef]
- Chobotow, J.; Strachecka, A. Morphology and function of insect fat bodies taking into account Apis mellifera L. honey bees. Med. Weter 2013, 69, 712–715. [Google Scholar]
- Załuski, R.; Bittarello, A.C.; Vieira, J.C.S.; Braga, C.P.; de Magalhaes Padilha, P.; da Silva Fernandes, M.; de Souza Bovi, T.; de Oliveira, O.R. Modification of the head proteome of nurse honey bees (Apis mellifera) exposed to field-relevant doses of pesticides. Sci. Rep. 2020, 10, 2190. [Google Scholar] [CrossRef]
- Gätschenberger, H.; Azzami, K.; Tautz, J.; Beier, H. Antibacterial immune competence of honey bees (Apis mellifera) is adapted to different life stages and environmental risks. PLoS ONE 2013, 8, e66415. [Google Scholar] [CrossRef] [PubMed]
- Hulbert, A.J.; Pamplona, R.; Buffenstein, R.; Buttemer, W.A. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol. Rev. 2007, 87, 1175–1213. [Google Scholar] [CrossRef] [PubMed]
- Cocchi, M.; Lercker, G.; Frega, N.G.; Gabrielli, F.; Quaranta, M. Is a low concentration of linoleic acid related to the extended longevity of the queen honey bee? Prog. Nutr. 2019, 21, 729–734. [Google Scholar]
- Park, H.G.; Lee, K.S.; Kim, B.Y.; Yoon, H.J.; Choi, S.Y.; Lee, K.Y.; Wan, H.; Li, J.; Jin, B.R. Honey bee (Apis cerana) vitellogenin acts as an antimicrobial and antioxidant agent in the body and venom. Dev. Comp. Immunol. 2018, 85, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Corona, M.; Velarde, R.A.; Remolina, S.; Moran-Lauter, A.; Wang, Y.; Kimberly, A.; Hughes, A.K.; Robinson, G.E. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. USA 2007, 104, 7128–7133. [Google Scholar] [CrossRef]
- Santos, D.E.; Souza, A.O.; Tibério, G.J.; Alberici, L.C.; Hartfelder, K. Differential expression of antioxidant system genes in honey bee (Apis mellifera L.) caste development mitigates ROS-mediated oxidative damage in queen larvae. Genet. Mol. Biol. 2020, 43, e20200173. [Google Scholar] [CrossRef]
- Mardani-Talaee, M.; Rahimi, V.; Zibaee, A. Effects of host plants on digestive enzymatic activities and some components involved in intermediary metabolism of Chrysodeixis chalcites (Lepidoptera: Noctuidae). J. Entomol. Acarol. Res. 2014, 46, 96–101. [Google Scholar] [CrossRef]
- Skowronek, P.; Wójcik, Ł.; Strachecka, A. Cannabis extract has a positive–immunostimulating effect through proteolytic system and metabolic compounds of honey bee (Apis mellifera) workers. Animals 2021, 11, 2190. [Google Scholar] [CrossRef]
| Queen | Rebel | Normal Workers | Queen | Rebel | Normal Workers | Queen | Rebel | Normal Workers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| Acidic Proteases (U/mg) | Neutral Proteases (U/mg) | Alkaline Proteases (U/mg) | |||||||||||||||||
| tissues | sternit | 0.61 | 0.05 | 1.00 | 0.06 | 1.25 | 0.08 | 0.35 | 0.04 | 0.44 | 0.03 | 1.26 | 0.08 | 2.86 | 0.33 | 2.96 | 0.14 | 3.37 | 0.19 |
| tergit 3 | 3.25 | 0.33 | 3.55 | 0.16 | 4.34 | 0.18 | 3.07 | 0.25 | 3.42 | 0.20 | 4.19 | 0.19 | 3.26 | 0.21 | 3.51 | 0.21 | 4.42 | 0.22 | |
| tergit 5 | 1.43 | 0.10 | 1.34 | 0.04 | 1.17 | 0.05 | 2.44 | 0.16 | 2.30 | 0.10 | 1.99 | 0.10 | 2.51 | 0.14 | 2.40 | 0.06 | 2.19 | 0.08 | |
| hemolymph | 0.75 | 0.07 | 0.84 | 0.03 | 1.06 | 0.04 | 0.76 | 0.08 | 0.92 | 0.04 | 2.30 | 0.14 | 0.96 | 0.06 | 0.82 | 0.03 | 0.06 | 0.00 | |
| Three-Way Anova | colony | F(2,684) = 1.50; p = 0.492 | F(2,684) = 7.75; p = 0.80 | F(2,684) = 0.41; p = 0.695 | |||||||||||||||
| phenotype | F(2,4) = 123.98; p = 0.0003 | F(2,4) = 555.98; p = 0.000 | F(2,4) = 8.18; p = 0.038 | ||||||||||||||||
| tissue | F(3,6) = 4019.96; p = 0.000 | F(3,6) = 5447.52; p = 0.000 | F(3,6) = 1285.66; p = 0.000 | ||||||||||||||||
| colony * phenotype | F(4,12) = 0.89; p = 0.501 | F(4,12) = 0.68; p = 0.618 | F(4,12) = 0.70; p = 0.607 | ||||||||||||||||
| colony * tissue | F(6,12) = 0.74; p = 0.631 | F(6,12) = 0.459; p = 0.826 | F(6,12) = 1.69; p = 0.206 | ||||||||||||||||
| phenotype * tissue | F(6,12) = 47.82; p = 0.000 | F(6,12) = 113.15; p = 0.000 | F(6,12) = 92.98; p = 0.000 | ||||||||||||||||
| colony * phenotype * tissue | F(12,684) = 8.10; p = 0.000 | F(12,684) = 7.031; p = 0.000 | F(12,684) = 6.34; p = 0.000 | ||||||||||||||||
| acidic protease inhibitors (U/mg) | neutral protease inhibitors (U/mg) | alkaline protease inhibitors (U/mg) | |||||||||||||||||
| tissues | sternit | 0 | 0 | 0.03 | 0.00 | 0.25 | 0.03 | 0 | 0 | 0.02 | 0.00 | 0.13 | 0.02 | 0 | 0 | 0.02 | 0.02 | 0.24 | 0.25 |
| tergit 3 | 0 | 0 | 0.15 | 0.02 | 0.54 | 0.03 | 0 | 0 | 0.01 | 0.00 | 0.24 | 0.03 | 0 | 0 | 0.03 | 0.04 | 0.34 | 0.35 | |
| tergit 5 | 0 | 0 | 0.14 | 0.02 | 0.33 | 0.02 | 0 | 0 | 0.01 | 0.00 | 0.14 | 0.02 | 0 | 0 | 0.03 | 0.03 | 0.33 | 0.34 | |
| hemolymph | 0 | 0 | 3.78 | 0.29 | 3.21 | 0.15 | 0 | 0 | 2.89 | 0.11 | 2.34 | 0.14 | 5.26 | 0.47 | 3.92 | 0.12 | 3.55 | 0.17 | |
| Three-Way Anova | colony | F(2,456) = 1.03; p = 0.675 | F(2,456) = 3.30; p = 0.675 | F(2,456) = 0.639; p = 0.608 | |||||||||||||||
| phenotype | F(1,2) = 8.93; p = 0.096 | F(1,2) = 4.0; p = 0.184 | F(2,4) = 108.83; p = 0.001 | ||||||||||||||||
| tissue | F(3,6) = 11,659.16; p = 0.000 | F(3,6) = 11,257.50; p = 0.000 | F(3,6) = 71,825.71; p = 0.000 | ||||||||||||||||
| colony * phenotype | F(2,6) = 1.03; p = 0.412 | F(2,6) = 0.959; p = 0.435 | F(2,6) = 2.90; p = 0.118 | ||||||||||||||||
| colony * tissue | F(6,12) = 0.56; p = 0.752 | F(6,12) = 0.105; p = 0.992 | F(6,12) = 0.256; p = 0.939 | ||||||||||||||||
| phenotype * tissue | F(3,6) = 113.73; p = 0.000 | F(3,6) = 241.70; p = 0.000 | F(3,6) = 224.79; p = 0.000 | ||||||||||||||||
| colony * phenotype * tissue | F(6,456) = 3.82; p = 0.001 | F(6,456) = 3.761; p = 0.001 | F(6,570) = 1.19; p = 0.304 | ||||||||||||||||
| Queen | Rebel | Normal Workers | Queen | Rebel | Normal Workers | Queen | Rebel | Normal Workers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| SOD (U/mg) | CAT (U/mg) | TAC (mM of Trolox) | |||||||||||||||||
| tissues | sternit | 0.44 | 0.03 | 0.48 | 0.04 | 0.75 | 0.03 | 1.21 | 0.09 | 1.16 | 0.02 | 2.28 | 0.24 | 1.26 | 0.12 | 1.48 | 0.04 | 3.26 | 0.19 |
| tergit 3 | 0.44 | 0.03 | 0.49 | 0.03 | 0.55 | 0.03 | 0.84 | 0.05 | 1.03 | 0.10 | 1.16 | 0.04 | 11.78 | 0.98 | 10.22 | 0.40 | 9.49 | 0.39 | |
| tergit 5 | 0.44 | 0.03 | 0.46 | 0.03 | 0.64 | 0.03 | 1.07 | 0.13 | 1.20 | 0.14 | 1.62 | 0.05 | 1.55 | 0.20 | 3.12 | 0.22 | 3.47 | 0.23 | |
| hemolymph | 0.44 | 0.03 | 0.60 | 0.05 | 1.15 | 0.05 | 7.41 | 0.45 | 7.11 | 0.18 | 5.38 | 0.23 | 3.61 | 0.24 | 48.07 | 0.65 | 45.54 | 0.51 | |
| Three-Way Anova | colony | F(2,684) = 0.12; p = 0.886 | F(2,684) = 18.43; p = 0.695 | F(2,684) = 0.852; p = 0.740 | |||||||||||||||
| phenotype | F(2,4) = 4786.06; p = 0.000 | F(2,4) = 0.18; p = 0.840 | F(2,4) = 5486.0; p = 0.000 | ||||||||||||||||
| tissue | F(3,6) = 1198.62; p = 0.000 | F(3,6) = 25,638.1; p = 0.000 | F(3,6) = 21,196.0; p = 0.000 | ||||||||||||||||
| colony * phenotype | F(4,12) = 2.56; p=0.092 | F(4,12) = 0.765; p = 0.56 | F(4,12) = 0.7; p = 0.632 | ||||||||||||||||
| colony * tissue | F(6,12) = 2.89; p = 0.055 | F(6,12) = 0.28; p = 0.935 | F(6,12) = 0.6; p = 0.701 | ||||||||||||||||
| phenotype * tissue | F(6,12) = 2002.14; p = 0.000 | F(6,12) = 189.02; p = 0.000 | F(6,12) = 3445.0; p = 0.000 | ||||||||||||||||
| colony * phenotype * tissue | F(12,684) = 0.48; p = 0.927 | F(12,684) = 5.99; p = 0.000 | F(12,684) = 23.5; p = 0.000 | ||||||||||||||||
| Queen | Rebel | Normal Workers | Queen | Rebel | Normal Workers | Queen | Rebel | Normal Workers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| AST (U/dm3) | ALT (U/dm3) | ALP (U/dm3) | |||||||||||||||||
| tissues | sternit | 25.4 | 0.36 | 22.3 | 0.46 | 21.7 | 0.39 | 30.3 | 0.53 | 25.3 | 0.63 | 24.4 | 0.47 | 7.5 | 0.24 | 6.29 | 0.43 | 5.44 | 0.26 |
| tergit 3 | 16.4 | 0.41 | 15.2 | 0.45 | 12.5 | 0.37 | 22.4 | 0.40 | 21.2 | 0.45 | 19.5 | 0.38 | 4.33 | 0.23 | 3.26 | 0.27 | 2.37 | 0.19 | |
| tergit 5 | 12.2 | 1.32 | 12 | 0.38 | 10.6 | 0.28 | 25.5 | 0.26 | 24.7 | 0.35 | 20.7 | 0.42 | 3.81 | 0.17 | 2.68 | 0.20 | 1.85 | 0.05 | |
| hemolymph | 26.4 | 0.37 | 23.5 | 0.30 | 22.4 | 0.49 | 28.5 | 0.45 | 25.9 | 0.40 | 25.5 | 0.39 | 6.53 | 0.24 | 5.52 | 0.23 | 5.44 | 0.30 | |
| Three-Way Anova | colony | F(2,684) = 1.4; p = 0.537 | F(2,684) = 0.4; p = 0.2 | F(2,684) = 0.9; p = 0.469 | |||||||||||||||
| phenotype | F(2,4) = 1787.9; p = 0.000 | F(2,4) = 7450; p = 0.000 | F(2,4) = 1261.26; p = 0.000 | ||||||||||||||||
| tissue | F(3,6) = 64,287.6; p = 0.000 | F(3,6) = 12,184; p = 0.000 | F(3,6) = 7220.73; p = 0.000 | ||||||||||||||||
| colony * phenotype | F(4,12) = 1.3; p = 0.325 | F(4,12) = 0.0; p = 0.773 | F(4,12) = 2.83; p = 0.073 | ||||||||||||||||
| colony * tissue | F(6,12) = 0.4; p = 0.879 | F(6,12) = 0.0; p = 0.902 | F(6,12) =1.51; p = 0.255 | ||||||||||||||||
| phenotype * tissue | F(6,12) = 146.3; p = 0.000 | F(6,12) = 254; p = 0.000 | F(6,12) = 67.31; p = 0.000 | ||||||||||||||||
| colony * phenotype * tissue | F(12,684) = 1.0; p = 0.461 | F(12,684) = 2; p = 0.07 | F(12,684) = 0.87; p = 0.575 | ||||||||||||||||
| Queen | Rebel | Normal Workers | |||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| Ovariole number | 199.55 | 25.36 | 12.37 | 1.84 | 5.08 | 1.07 | |
| Two-Way Anova | colony | F(2,4) = 1.002; p = 0.444 | |||||
| phenotype | F(2,4) = 1442.48; p = 0.000 | ||||||
| colony * phenotype | F(4,171) = 2.458; p = 0.047 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strachecka, A.; Kuszewska, K.; Olszewski, K.; Skowronek, P.; Grzybek, M.; Grabowski, M.; Paleolog, J.; Woyciechowski, M. Activities of Antioxidant and Proteolytic Systems and Biomarkers in the Fat Body and Hemolymph of Young Apis mellifera Females. Animals 2022, 12, 1121. https://doi.org/10.3390/ani12091121
Strachecka A, Kuszewska K, Olszewski K, Skowronek P, Grzybek M, Grabowski M, Paleolog J, Woyciechowski M. Activities of Antioxidant and Proteolytic Systems and Biomarkers in the Fat Body and Hemolymph of Young Apis mellifera Females. Animals. 2022; 12(9):1121. https://doi.org/10.3390/ani12091121
Chicago/Turabian StyleStrachecka, Aneta, Karolina Kuszewska, Krzysztof Olszewski, Patrycja Skowronek, Maciej Grzybek, Marcin Grabowski, Jerzy Paleolog, and Michał Woyciechowski. 2022. "Activities of Antioxidant and Proteolytic Systems and Biomarkers in the Fat Body and Hemolymph of Young Apis mellifera Females" Animals 12, no. 9: 1121. https://doi.org/10.3390/ani12091121
APA StyleStrachecka, A., Kuszewska, K., Olszewski, K., Skowronek, P., Grzybek, M., Grabowski, M., Paleolog, J., & Woyciechowski, M. (2022). Activities of Antioxidant and Proteolytic Systems and Biomarkers in the Fat Body and Hemolymph of Young Apis mellifera Females. Animals, 12(9), 1121. https://doi.org/10.3390/ani12091121








