Simple Summary
The coat color of dromedary is usually uniform and varies from black to white. We identified 9 significant SNPs associated with white color, and the 13 significant SNPs associated with black color using genotyping-by-sequencing (GBS). Among candidate genes, SNAI1 that interacts with MCIR, ASIP and KIT genes plays a key role in the melanin biosynthetic and pigmentation biological process and melanogenesis biological pathway.
Abstract
The coat color of dromedary is usually uniform and varies from black to white, although dark- to light-brown colors are the most common phenotypes. This project was designed to gain knowledge on novel color-related variants using genotyping-by-sequencing (GBS). The association between the SNPs and coat color was tested using MLM (mixed linear models) with kinship matrix. Three GWAS models including white color vs. non-white color, black vs. non-black color, and light-brown vs. dark-brown color were performed. There were no distinct genetic clusters detected based on the color phenotypes. However, admixture occurred among all individuals of the four different coat color groups. We identified nine significant SNPs associated with white color after Bonferroni correction, located close to ANKRD26, GNB1, TSPYL4, TEKT5, DEXI, CIITA, TVP23B, CLEC16A, TMPRSS13, FXYD6, MPZL3, ANKRD26, HFM1, CDC7, TGFBR3, and HACE1 genes in neighboring flanking regions. The 13 significant SNPs associated with black color and the candidate genes were: CAPN7, CHRM4, CIITA, CLEC16A, COL4A4, COL6A6, CREB3L1, DEXI, DGKZ, DGKZ, EAF1, HDLBP, INPP5F, MCMBP, MDK, SEC23IP, SNAI1, TBX15, TEKT5, TMEM189, trpS, TSPYL4, TVP23B, and UBE2V1. The SNAI1 gene interacted with MCIR, ASIP and KIT genes. These genes play a key role in the melanin biosynthetic and pigmentation biological process and melanogenesis biological pathway. Further research using a larger sample size and pedigree data will allow confirmation of associated SNPs and the identified candidate genes.
1. Introduction
There is a large diversity of coat colors and patterns in livestock, which is important for an easy and rapid discrimination among breeds, species, or ecotypes [1]. Often this trait has been artificially selected, as the value of natural-colored wool depends on the fiber quality and coat color of the animal (e.g., merino sheep, cashmere goat, alpaca) [2]. Additionally, selective breeding in farm animals has been traditionally based on morphological characters based on coat coloration [3]. However, natural selection for adaptation to environment also acts on coat color variation [4], e.g., black-coated animals attract heat more than others in harsh deserts, supporting resistance to high thermal differences [5]. The color of dromedaries is uniform and varies from white (e.g., Wodh) to black (e.g., Magaheem). While few West African camels are spotted [6], dark- to light-brown colors are the most common phenotypes. Based on these colors, Saudi Arabian camels were divided into four breeds: white (Magateer), brown (Al Homr and Al Sofr), and black (Magaheem) [7]. Heredity of coat color is a favorite area of genetics research because of its highly visible nature [8]. The biogenesis of pigment granules is affected by multiple genes [9], and more than 50 genes have been reported affecting coat color in mammals [10]. Well-defined genes for influencing coat color are the agouti signaling protein (ASIP or Agouti), melanocyte-stimulating hormone receptor (MC1R), and proto-oncogene receptor tyrosine kinase (KIT) [11,12,13,14,15,16,17,18,19,20,21,22]. MC1R and ASIP affect the ratio of eumelanin and pheomelanin distribution in mammals and particularly in domesticated animals [1,3,17]. Eumelanin is responsible for black, brown, or grey and pheomelanin for red, yellow or cream color, respectively [23]. In camelids, a few projects have already studied coat color [1,6,14,24,25,26]. In alpaca, it has shown that MC1R and ASIP genes are the main candidates for coat color [25]. Three SNPs located on MC1R (901 (C/T)), and TYRP1 (113 (C/T) and 200 (C/T)) were identified in Pakistani dromedaries [24]. ASIP and MCIR were recognized as candidate genes for black and white coat colors, respectively, in dromedaries [1]. The protein MC1R plays a key role in pigmentation and determines coat color via dark eumelanin or light pheomelanin [1]. α-MSH as a melanocyte stimulating hormone connected to the MC1R receptor to produce eumelanin. It seems that ASIP gene regulates expression of MC1R gene and reduces eumelanin, causing the brown coat color in dromedary [1]. Although most dromedary populations are brown, its intensity differs from white to black color in different breeds and populations of dromedaries, which can be due to epistatic interactions with other gene products besides those from MC1R and ASIP genes [1]. With next generation sequencing platforms and genome-wide association studies it is becoming more and more efficient to study the mechanism of pigmentation. In Iran, there are about 140,000 dromedaries [27]. They are usually light- and dark-brown color.
We designed this project to gain knowledge on novel color-related variants, and we applied a genome-wide association study approach using genotyping-by-sequencing (GBS). We aimed at identifying novel SNPs associated with pigmentation in dromedaries. The ultimate goal of the study was to identify proteins associated with the various shades of pigmentation observed in this livestock species.
2. Materials and Methods
The work was approved by ASRI’s Animal Ethics Committee (the number ASRI-34–64-1357–005-970,180). The blood samples were gathered from 96 dromedaries in Iran central desert including the following: light-brown (n = 42), dark-brown (n = 35), black (n = 9), and white (n = 10). DNA Extraction was performed by the modified salt precipitation method [28]; after elution, quality was checked by spectrophotometry. Finally, DNA samples were adjusted to a concentration of 50 ng/µL for subsequent steps. EcoR1 and HinF1 enzymes were used to genotyping-by-sequencing, based on paired-end (150 bp) sequencing. GBS library was composed in three steps: cutting the DNA using Restriction enzymes, attaching adaptor to the cut DNA, and amplifying DNA Molecules using DNA polymerase. This project was carried out using the Illumina HiSeq 2000 platform in Persian Bayangene Research and Training Center (Shiraz, Iran). Trimming of adapters and quality control of read pairs (base Qphred ≤20) were performed using bcl2fastq V2.20 and fastQC V1.0.0 tools. Mapping of sequence reads (GCA_000803125.3) [29], PCR duplicates detection, and SNP calling were executed using the BWA-MEM algorithm of Burrows–Wheeler Aligner (BWA) [30], Picard tools [31], SAMtools [32], and GATK, respectively. Variants with MAF <0.05 and Call Rate < 0.95 were removed. The association between the SNPs and coat color were estimated by MLM (mixed linear models) with kinship matrix [32]. In this project, the regions of sampling were included as fixed effect. We performed the case–control GWAS in three sets. In set I, we treated all the white colors as case groups (n = 10) and non-white appearance as controls (n = 86). In set II, the black colors were treated as case groups (n = 9) and non-black appearance as controls (n = 87). In the remaining set, we treated light brown (n = 42) as case groups and dark brown color as controls (n = 35).
The statistical model was as follows:
Y = αX + UZ + e
- Y: coat color
- α: SNP effects
- U: kinship background effects
- e: residual effects
- X and Z: incidence matrix relating the individuals to fixed marker effects α and random group effects u, respectively.
We used Bonferroni-corrected threshold (-log p value > 4.16), which were defined as 0.05/N (N is the number of tested SNPs). The candidate genes were detected by tracing the associated SNPs in NCBI (GCA_000803125.3) [29], located either within the exon/intron of a gene or within a flanking region of 100 kb up- and downstream. vcfR, poppr, ape, and RColorBrewer packages in R V4.1.2 [33,34,35,36] were used for K-means clustering and discriminant analysis of principal components (DAPC) and TASSEL V5.0 [37] for generation of Manhattan and q–q plot. PPI enrichment analysis was performed by STRING database [38].
3. Results
3.1. Summary Statistic of SNPs, Linkage Disequilibrium, Population Structure, and Kinship Analyses
The number of 41,897 variants were discovered. A total of 14,522 SNPs remained after quality control, so that 256 monomorphic markers and 27,375 markers with MAF < 0.05 were removed. Principle component analysis (PCA) showed that the camels of five different regions of Iran central desert were homogenous (Figure 1A). As there were no distinctive clusters detected based on sampling regions, admixture among all individuals of the four different coat colors must have occurred (Figure 1B).
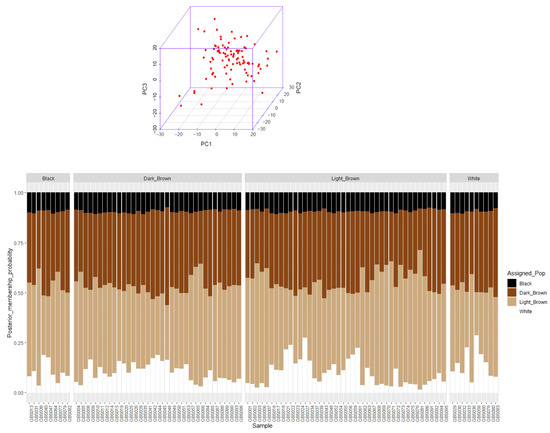
Figure 1.
Principal component analysis of 96 dromedary camels of Iran central desert using 14,522 SNPs in five regions in above figure (A). Admixture of 96 dromedary camels among four coat colors: light-brown, dark-brown, black, and white, where the x-axis shows sample number and the y-axis show the probability of each camel belongs to coat color (color = K) in bottom figure (B).
3.2. Genome-Wide Association Study
GWAS results for the set I and set II are presented in Figure 2 and Figure 3, respectively. We identified nine significant SNPs associated with white color after Bonferroni correction, located in neighboring flanking regions close to ANKRD26, GNB1, TSPYL4, TEKT5, DEXI, CIITA, TVP23B, CLEC16A, TMPRSS13, FXYD6, MPZL3, HFM1, CDC7, TGFBR3, and HACE1 genes in neighboring flanking regions (Table 1 and Figure 2). The 13 significant SNPs associated with black color and their candidate genes were: CAPN7, CHRM4, CIITA, CLEC16A, COL4A4, COL6A6, CREB3L1, DEXI, DGKZ, DGKZ, EAF1, HDLBP, INPP5F, MCMBP, MDK, SEC23IP, SNAI1, TBX15, TEKT5, TMEM189, trpS, TSPYL4, TVP23B, and UBE2V1. We did not find any SNPs associated with light- and dark-brown color in this study (Table 1 and Figure 3). SNP Chr19_10157184 located close to SNAI1 gene that the result of PPI enrichment analysis by using STRING database indicated SNAI1 gene interacts with MCIR, ASIP and KIT genes. Network analysis of SNAI1, MCIR, ASIP, and KIT genes showed that the number of nodes is eight, the number of edges is 17, the average degree of node is 4.25, the clustering coefficient is 0.86 and it is significant based on p_value < 0.001.
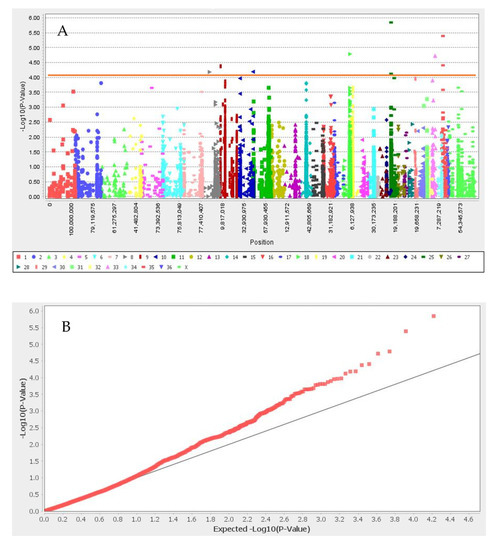
Figure 2.
Manhattan plot (A) and q–q plot (B) of coat color for white color. Red line in Manhattan plot is Bonferroni threshold. The chromosomal position of each SNP was displayed in different colors in Manhattan plot. Red and gray line in the q–q plots represent the −log p-value of the entire study and expected value, respectively.
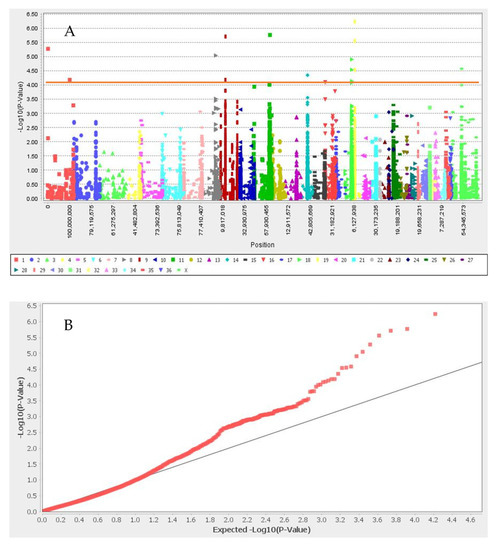
Figure 3.
Manhattan plot (A) and q–q plot (B) of coat color for black color. Red line in Manhattan plot is Bonferroni threshold. The chromosomal position of each SNP was displayed in different colors in Manhattan plot. Red and gray line in the q–q plots represent the −log p-value of the entire study and expected value, respectively.

Table 1.
The associated SNPs in three case–control GWAS models of coat color in dromedaries.
4. Discussion
In this research, genome-wide association analysis with 15K SNPs has been per-formed with an attempt to clarify the SNPs associated with various coat colors exhibited in dromedaries. Most dromedaries from Iran central desert included in this study had brown coat colors. We identified 22 markers that were significantly associated with coat color in dromedaries; thus, nine SNPs included the following: Chr25_73462, Chr35_5648539, Chr18_29916170, Chr33_12297059, Chr35_5696438, Chr9_1807083, Chr10_74907708, Chr8_31826040, and Chr25_505194 related with white color; 13 SNPs included the following: Chr19_10157184, Chr11_74286851, Chr9_22746017, Chr19_10612243, Chr1_2037209, Chr8_59919441, Chr18_29898490, Chrx_46816486, Chr18_29898527, Chr19_10612244, Chr14_30853969, Chr9_22614201, and Chr1_101414918 with black color. The marker called Chr35_5648539, associated with white color, located near to GNB1 gene and functional profiling of this gene, suggested phototransduction with KEGG: 04744 (p-value = 2.39 × 10−2). The marker called Chr1_2037209, associated with black color, located near to COL4A4, COL6A6 genes and collagen type IV trimer term with GO:0005587 (p-value = 2.90 × 10−2), was detected for these genes. The marker called Chr19_10157184, located close to SNAI1, interacted with MCIR, ASIP and KIT genes, which together play a key role in the melanin biosynthetic and pigmentation biological process and melanogenesis biological pathway. The black and brown coat colors are probably caused by the ASIP gene in dromedaries [1], while the white coat color is likely regulated by the MC1R gene in Wodh dromedaries [1]. Heterozygosity of c.901C >T in MC1R as well as the homozygous genotype of ASIP exon 2 are responsible for the white color phenotype. Likewise, several researchers have reported MC1R, ASIP genes related to coat color in cattle, goats, and sheep [39,40,41,42,43,44,45,46]. Holl et al. (2017) found that a mutation in the KIT gene is associated with white-spotted phenotypes in the dromedary [22]. TYR is another basic gene that regulates pigmentation [47] and disorders in this gene cause the albino phenotype in mammals and chickens [48]. Genetic diversity of indigenous dromedaries is at risk in Iran [49], their genetic diversity needs to be conserved. One of approaches to prediction of animal breed is coat color, so that morphological selection in domestic animals based on color determines the breed and attribution [17]. The reported SNPs in this study represent the candidate polymorphisms associated with pigmentation in dromedaries and can be used in future genetic studies. However, whole genome sequencing, pedigree, and more phenotype data will be required to accurate survey of coat color in dromedaries.
5. Conclusions
Fiber color is important in textile industry [50], and the Asian camel farmers show high interest for breeding white and black dromedaries [1]. Genetic analyses indicated that several loci are involved in coat color in dromedaries that can be used in selective breeding or towards identification of purebred animals. It would be very interesting to survey indigenous dromedaries regarding their color evolution, using a larger sample size and pedigree which should include many populations and breeds. In addition, more detailed studies are needed in order to understand the associated SNPs and candidate genes identified in this project.
Author Contributions
Conceptualization, M.B.S., J.Z.H., S.E., M.H.B., A.B., B.M.N., M.Z., N.A. and N.S.; methodology, M.B.S., J.Z.H., M.H.B., M.A.F., M.S., A.T.S., A.T. and A.S.N.; software, M.B.S., Z.R. and M.A.F.; formal analysis, M.B.S.; investigation, M.B.S. and J.Z.H.; writing—original draft preparation, M.B.S.; writing—review and editing, S.E., M.H.B., P.A.B., A.F. and M.A.F.; visualization, M.B.S.; supervision, M.H.B., S.E., M.A.F. and P.A.B. All authors have read and agreed to the published version of the manuscript.
Funding
PB acknowledges funding from the Austrian Science Fund (FWF): P29623-B25. This research was jointly funded by Animal Science Research Institute of Iran (ASRI), Animal Breeding Center of Iran, and Yazd Agricultural and Natural Resources Research and Education Center, grant number 34-64-1357-005-970180.
Institutional Review Board Statement
The work was approved by ASRI’s Animal Ethics Committee (the number ASRI-34–64-1357–005-970,180).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and during the current study are available in the [dryad] repository, [https://datadryad.org/stash].(accessed on 22 April 2022).
Acknowledgments
We would like to thank the camel farmers in Yazd province for providing samples. Open Access Funding by the Austrian Science Fund (FWF).
Conflicts of Interest
The authors declare they have no competing interests.
References
- Almathen, F.; Elbir, H.; Bahbahani, H.; Mwacharo, J.; Hanotte, O. Polymorphisms in MC1R and ASIP Genes are Associated with Coat Color Variation in the Arabian Camel. J. Hered. 2018, 109, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.J.; Horst, P.; Kleinheisterkamp, H.H. The importance of coat colour and coat type as indicators of productive adaptability of beef cattle in a subtropical environment. Trop. Anim. Prod. 1982, 7, 296–304. [Google Scholar]
- Giantsis, I.A.; Antonopoulou, D.; Dekolis, N.; Zaralis, K.; Avdi, M. Origin, demographics, inbreeding, phylogenetics, and phenogenetics of Karamaniko breed, a major common ancestor of the autochthonous Greek sheep. Trop. Anim. Health Prod. 2022, 54, 73. [Google Scholar] [CrossRef] [PubMed]
- Finch, V.A.; Bennett, I.L.; Holmes, C.R. Coat colour in cattle: Effect on thermal balance, behaviour and growth, and relationship with coat type. J. Agric. Sci. 1984, 102, 141–147. [Google Scholar] [CrossRef]
- Ward, J.M.; Blount, J.D.; Ruxton, G.D.; Houston, D.C. The adaptive significance of dark plumage for birds in desert environments. Ardea 2002, 90, 311–323. [Google Scholar]
- Traoré, B.; Moula, N.; Toure, A.; Ouologuem, B.; Leroy, P.; Antoine-Moussiaux, N. Characterisation of camel breeding practices in the Ansongo Region, Mali. Trop. Anim. Health Prod. 2014, 46, 1303–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Swailem, A.M.; Al-Busadah, K.A.; Shehata, M.M.; Al-Anazi, I.O.; Askari, E. Classification of Saudi Arabian camel (Camelus dromedarius) subtypes based on RAPD technique. J. Food Agric. Environ. 2007, 5, 143–148. [Google Scholar]
- Hussein, Y.A.; Al-Eknah, M.M.; Al-Shami, S.A.; Mandour, M.A.; Fouda, T.A. Coat color breed variation in blood constituents among indigenous Saudi Arabia camel strains. Mansoura Vet. Med. J. 2012, 14, 191–204. [Google Scholar]
- Sturm, A.R.; Teasdale, R.D.; Box, N.F. Human pigmentation genes: Identification, structure and consequences of polymorphic variation. Gene 2001, 277, 49–62. [Google Scholar] [CrossRef]
- Cieslak, M.; Reissmann, M.; Hofreiter, M.; Ludwig, A. Colours of domestication. Biol. Rev. 2011, 86, 885–899. [Google Scholar] [CrossRef]
- Geissler, E.N.; Ryan, M.A.; Housman, D.E. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell 1988, 55, 185–192. [Google Scholar] [CrossRef]
- Marklund, S.; Kijas, J.; Rodriguez-Martinez, H.; Rönnstrand, L.; Funa, K.; Moller, M.; Lange, D.; Edfors-Lilja, I.; Andersson, L. Molecular Basis for the Dominant White Phenotype in the Domestic Pig. Genome Res. 1998, 8, 826–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pielberg, G.; Olsson, C.; Syvanen, A.C.; Andersson, L. Unexpectedly high allelic diversity at the KIT locus causing dominant white color in the domestic pig. Genetics 2002, 160, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Haase, B.; Brooks, S.A.; Tozaki, T.; Burger, D.; Poncet, P.A.; Rieder, S.; Hasegawa, T.; Penedo, C.; Leeb, T. Seven novel KIT mutations in horses with white coat colour phenotypes. Anim. Genet. 2009, 40, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Haase, B.; Jagannathan, V.; Rieder, S.; Leeb, T. A novel KIT variant in an Icelandic horse with white-spotted coat colour. Anim. Genet. 2015, 46, 466. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Dall’Olio, S.; González, E.G.; Finocchiaro, R.; Davoli, R.; Russo, V.; Portolano, B. Missense and nonsense mutations in melanocortin 1 receptor (MC1R) gene of different goat breeds: Association with red and black coat colour phenotypes but with unexpected evidences. BMC Genet. 2009, 10, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatis, C.; Giannoulis, T.; Galliopoulou, E.; Billinis, C.; Mamuris, Z. Genetic analysis of melanocortin 1 receptor gene in endangered Greek sheep breeds. Small Rumin. Res. 2017, 157, 54–56. [Google Scholar] [CrossRef]
- Fontanesi, L.; D’Alessandro, E.; Scotti, E.; Liotta, L.; Crovetti, A.; Chiofalo, V.; Russo, V. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Anim. Genet. 2010, 41, 478–492. [Google Scholar] [CrossRef]
- Wong, A.K.; Ruhe, A.L.; Robertson, K.R.; Loew, E.R.; Williams, D.C.; Neff, M.W. Ade novomutation inKITcauses white spotting in a subpopulation of German Shepherd dogs. Anim. Genet. 2012, 44, 305–310. [Google Scholar] [CrossRef]
- David, V.A.; Menotti-Raymond, M.; Wallace, A.C.; Roelke, M.; Kehler, J.; Leighty, R.; Eizirik, E.; Hannah, S.S.; Nelson, G.; Schäffer, A.A.; et al. Endogenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3: Genes Genomes Genet. 2014, 4, 1881–1891. [Google Scholar] [CrossRef] [Green Version]
- Dürig, N.; Jude, R.; Holl, H.; Brooks, S.A.; Lafayette, C.; Jagannathan, V.; Leeb, T. Whole genome sequencing reveals a novel deletion variant in the KIT gene in horses with white spotted coat colour phenotypes. Anim. Genet. 2017, 48, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Holl, H.; Isaza, R.; Mohamoud, Y.; Ahmed, A.; Almathen, F.; Youcef, C.; Gaouar, S.; Antczak, D.F.; Brooks, S. A Frameshift Mutation in KIT is Associated with White Spotting in the Arabian Camel. Genes. 2017, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Searl, A.G. Comparative Genetics of Coat Colour in Mammals; Logos: London, UK, 1968. [Google Scholar]
- Qureshi, A.S.; Shah, M.G.; Reiβmann, M.; Schwartz, H.J. Differentiation of six Pakistani camel breeds by molecular genetics analysis. In Proceedings of the ICAR Satellite Meeting on Camelid Reproduction, Budapest, Hungary, 12–13 July 2008; pp. 61–65. [Google Scholar]
- Cransberg, R.; Munyard, K.A. Polymorphisms detected in the tyrosinase and matp (slc45a2) genes did not explain coat colour dilution in a sample of Alpaca (Vicugna pacos). Small Rumin. Res. 2011, 95, 92–96. [Google Scholar] [CrossRef]
- Chandramohan, B.; Renieri, C.; La Manna, V.; La Terza, A. The alpaca agouti gene: Genomic locus, transcripts and causative mutations of eumelanic and pheomelanic coat color. Gene 2013, 521, 303–310. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. 2019. Available online: http://faostat3.fao.org/download/Q/QC/E (accessed on 21 January 2015).
- Javanrouh, A.; Banabazi, M.H.; Esmaeilkhanian, S.; Amirinia, C.; Seyedabadi, H.R.; Emrani, H. Optimization on salting out method for DNA extraction from animal and poultry blood cells. In Proceedings of the 57th Annual Meeting Of The European Association For Animal Production, Antalya, Turkey, 17–20 September 2006. [Google Scholar]
- Elbers, J.P.; Rogers, M.F.; Perelman, P.L.; Proskuryakova, A.A.; Serdyukova, N.A.; Johnson, W.E.; Horin, P.; Corander, J.; Murphy, D.; Burger, P.A. Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol. Ecol. Resour. 2019, 19, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pembleton, L.W.; Cogan, N.O.I.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Knaus, B.J.; Grünwald, N.J. vcfr: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef]
- Kamvar, Z.; Tabima, J.; Grunwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuwirth, E.; Neuwirth, M.E. Package ‘RColorBrewer’; CRAN 2011-06-17 08:34:00. Apache License 2.0; R Core Team: Vienna, Austria, 2011. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Alshanbari, F.; Castaneda, C.; Juras, R.; Hillhouse, A.; Mendoza, M.N.; Gutiérrez, G.A.; De León, F.A.P.; Raudsepp, T. Comparative FISH-Mapping of MC1R, ASIP, and TYRP1 in New and Old World Camelids and Association Analysis With Coat Color Phenotypes in the Dromedary (Camelus dromedarius). Front. Genet. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Klungland, H.; Våge, D.I.; Gomez-Raya, L.; Adalsteinsson, S.; Lien, S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm. Genome 1995, 6, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Joerg, H.; Fries, H.R.; Meijerink, E.; Stranzinger, G.F. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm. Genome 1996, 7, 317–318. [Google Scholar] [CrossRef]
- Våge, D.I.; Klungland, H.; Lu, D.; Cone, R.D. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm. Genome 1999, 10, 39–43. [Google Scholar] [CrossRef]
- Switonski, M.; Mankowska, M.; Salamon, S. Family of melanocortin receptor (MCR) genes in mammals—mutations, polymorphisms and phenotypic effects. J. Appl. Genet. 2013, 54, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Adalsteinsson, S.; Sponenberg, D.P.; Alexieva, S.; Russel, A.J. Inheritance of goat coat colors. J. Hered. 1994, 85, 267–272. [Google Scholar] [CrossRef]
- Martin, P.M.; Palhière, I.; Ricard, A.; Tosser-Klopp, G.; Rupp, R. Genome Wide Association Study Identifies New Loci Associated with Undesired Coat Color Phenotypes in Saanen Goats. PLoS ONE 2016, 11, e0152426. [Google Scholar] [CrossRef]
- Norris, B.J.; Whan, V.A. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 2008, 18, 1282–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, M.G.; Reissmann, M.; Qureshi, A.S.; Schwartz, H.J. Sequencing and mutation screening in exon 1 of camel tyrosinase gene. The global food & product chain-dynamics, innovations, conflicts, strategies; Deutscher Tropentag: Hohenheim, Germany, 11–13 October 2005. [Google Scholar]
- Schmidtz, B.H.; Buchanan, F.C.; Plante, Y.; Schmutz, S.M. Linkage mapping of Tyrosinase gene to bovine chromosome 26. Anim. Genet. 2001, 32, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Bitaraf Sani, M.; Harofte, J.Z.; Bitaraf, A.; Esmaeilkhanian, S.; Banabazi, M.H.; Salim, N.; Teimoori, A.; Naderi, A.S.; Faghihi, M.A.; Burger, P.A.; et al. Genome-Wide Diversity, Population Structure and Demographic History of Dromedaries in the Central Desert of Iran. Genes 2020, 11, 599. [Google Scholar] [CrossRef] [PubMed]
- Morante, R.; Goyache, F.; Burgos, A.; Cervantes, I.; Pérez-Cabal, M.; Gutiérrez, J. Genetic improvement for alpaca fibre production in the Peruvian Altiplano: The Pacomarca experience. Anim. Genet. Resour. Inf. 2009, 45, 37–43. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).