Cryptic Diversity of the European Blind Mole Rat Nannospalax leucodon Species Complex: Implications for Conservation
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. The Genus Nannospalax
1.2. The Role of Chromosomal Changes in Reproductive Isolation
1.3. Molecular Analysis of N. leucodon Genetic Differentiation
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Data Collection and Sequence Analysis
2.4. Genetic Diversity and Phylogenetic Analyses
3. Results
3.1. 16S rRNA Gene Nucleotide Sequence Comparison
3.2. MT-CYTB Gene Nucleotide Sequence Comparison
4. Discussion
4.1. Delineation of 11 N. leucodon CFs
4.2. Chromosomal Speciation, Reproductive Isolation, and the Biological Species Concept
4.3. Major Factors of Threat and Implications for Conservation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Isaac, N.J.B.; Mallet, J.; Mace, G.M. Taxonomic Inflation: Its Influence on Macroecology and Conservation. Trends Ecol. Evol. 2004, 19, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zachos, F.E.; Lovari, S. Taxonomic inflation and the poverty of the Phylogenetic Species Concept—A reply to Gippoliti and Groves. Hystrix Ital. J. Mammal. 2013, 24, 142–144. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Dudash, M.R.; Eldridge, M.D.B.; Fenster, C.B.; Lacy, R.C.; Mendelson, J.R., III; Porton, I.J.; Ralls, K.; Ryder, O.A. Implications of different species concepts for conserving biodiversity. Biol. Conserv. 2012, 153, 25–31. [Google Scholar] [CrossRef]
- Savić, I.; Ćirović, D.; Bugarski-Stanojević, V. Exceptional Chromosomal Evolution and Cryptic Speciation of Blind Mole Rats Nannospalax leucodon (Spalacinae, Rodentia) from South-Eastern Europe. Genes 2017, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.D.; Nevo, E.; Han, L.J.; Levanon, E.Y.; Zhao, J.; Avivi, A.; Larkin, D.; Jiang, X.; Feranchuk, S.; Zhu, Y.; et al. Genome-wide adaptive complexes to undergound stresses in blind mole rats. Nat. Commun. 2014, 5, 3966. [Google Scholar] [CrossRef]
- Malik, A.; Korol, A.; Hübner, S.; Hernandez, A.G.; Thimmapuram, J.; Ali, S.; Glaser, F.; Paz, A.; Avivi, A.; Band, M. Transcriptome Sequencing of the Blind Subterranean Mole Rat, Spalax galili: Utility and Potential for the Discovery of Novel Evolutionary Patterns. PLoS ONE 2011, 6, e21227. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, H.; Hangmann, J.; Shams, I.; Avivi, A.; Hankeln, T. Molecular evolution of antioxidant and hypoxia response in long-lived, cancer-resistant blind mole rats: The Nrf2-Keap1 pathway. Gene 2015, 577, 293–298. [Google Scholar] [CrossRef]
- Azpurua, J.; Seluanov, A. Long-lived cancer resistant rodents as new model species for cancer research. Front. Genet. 2013, 3, 319. [Google Scholar] [CrossRef] [Green Version]
- Manov, I.; Hirsh, M.; Iancu, T.; Malik, A.; Sotnichenko, N.; Band, M.; Avivi, A.; Shams, I. Pronounced cancer resistence in a subterranean rodent, the blind mole-rat, Spalax: In vivo and in vitro evidence. BMC Biol. 2013, 11, 91. [Google Scholar] [CrossRef] [Green Version]
- Savić, I.R. Ecology of the mole rat Spalax leucodon Nordm, in Yugoslavia. Proc. Nat. Sci. Matica Srp. Novi Sad 1973, 44, 5–70, (In Serbian with English Summary). [Google Scholar]
- Avivi, A.; Oster, H.; Joel, A.; Beiles, A.; Albrecht, U.; Nevo, E. Circadian genes in a blind subterranean mammal. III. Molecular cloning and circadian regulation of cryptochrome genes in the blind subterranean mole rat, Spalax ehrenbergi superspecies. J. Biol. Rhythm. 2004, 19, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E. Mosaic Evolution of Subterranean Mammals: Regression, Progression and Global Convergence; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Arslan, A.; Kryštufek, B.; Matur, F.; Zima, J. Review of chromosome races in blind mole rats (Spalax and Nannospalax). Folia Zool. 2016, 65, 249–301. [Google Scholar] [CrossRef]
- Kankılıç, T.; Arslan, S.; Şeker, P.S.; Kankılıç, T.; Toyran, K.; Zima, J. A new chromosomal race (2n = 44) of Nannospalax xanthodon from Turkey (Mammalia: Rodentia). Zool. Middle East 2017, 63, 181–188. [Google Scholar] [CrossRef]
- Savić, I.; Soldatović, B. Karyotype Evolution and Taxonomy of the Genus Nannospalax Palmer, 1903, Mammalia, in Europe; Serbian Academy of Sciences and Arts, Separate Editions: Belgrade, Serbia, 1984; Volume 59, pp. 1–104. [Google Scholar]
- Savić, I.; Soldatović, B. Distribution range and evolution of chromosomal forms in the Spalacidae of the Balkan Peninsula and bordering regions. J. Biogeogr. 1979, 6, 363–374. [Google Scholar] [CrossRef]
- Kryštufek, B. Nannospalax leucodon (Nordmann, 1840). In The Atlas of European Mammals; Mitchell-Jones, A.J., Amori, G., Bogdanowicz, W., Kryštufek, B., Reijnders, P.J.H., Spitzenberger, F., Stubbe, M., Thissen, J.B.M., Vohralík, V., Zima, J., Eds.; Academic Press: London, UK, 1999; pp. 262–263. [Google Scholar]
- Wahrman, J.; Goitein, R.; Nevo, E. Mole rat Spalax: Evolutionary significance of chromosome variation. Science 1969, 164, 82–84. [Google Scholar] [CrossRef]
- Lyapunova, E.A.; Vorontsov, N.N.; Martynova, L. Cytological differentiation of burrowing mammals in the Palaearctic. In Symposium Theriologicum II; Kratochvíl, J., Obrtel, R., Eds.; Academia: Prague, Czech Republic, 1974; pp. 203–215. [Google Scholar]
- Vorontsov, N.N.; Martynova, L.N.; Fomicheva, I.I. An electrophoretic comparison of the blood proteins in mole rats of the fauna of the USSR (Spalacinae, Rodentia). Zool. Zh. 1977, 56, 1207–1215, (In Russian with English Summary). [Google Scholar]
- Savić, I.R. Familie Spalacidae Gray, 1821—Blindmäuse. In Handbuch der Säugetiere Europas; Niethammer, J., Krapp, F., Eds.; Akademische Verlagsgesellschaft: Wiesbaden, Germany, 1982; pp. 539–584. [Google Scholar]
- Kryštufek, B.; Amori, G. Nannospalax leucodon (amended version of 2008 assessment). In The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2017. [Google Scholar]
- Nevo, E.; Ivanitskaya, E.; Beiles, A. Adaptive Radiation of Blind Subterranean Mole Rats: Naming and Revisiting the Four Siblingspecies of the Spalax ehrenbergi Superspecies in Israel: Spalax galili (2n = 52), S. golani (2n = 54), S. carmeli (2n = 58) and S. judaei (2n = 60); Bachkhuys Publishers: Leiden, The Netherlands, 2001. [Google Scholar]
- Ivanitskaya, E.; Belyayev, A.; Nevo, E. Heterochromatin differentiation shows the pathways of karyotypic evolution in Israeli mole rats (Spalax, Spalacidae, Rodentia). Cytogenet. Genome Res. 2005, 111, 159–165. [Google Scholar] [CrossRef]
- Ivanitskaya, E.; Sözen, M.; Rashkovetsky, L.; Matur, F.; Nevo, E. Discrimination of 2n = 60 Spalax leucodon cytotypes (Spalacidae, Rodentia) in Turkey by means of classical and molecular cytogenetic techniques. Cytogenet. Genome Res. 2008, 122, 139–149. [Google Scholar] [CrossRef]
- Castiglia, R. Sympatric sister species in rodents are more chromosomally differentiated than allopatric ones: Implications for the role of chromosomal rearrangements in speciation. Mammal Rev. 2013, 44, 1–4. [Google Scholar] [CrossRef]
- Bulatova, N.S.; Biltueva, L.S.; Pavlova, S.V.; Zhdanova, N.S.; Zima, J. Chromosomal differentiation in the common shrew and related species. In Shrews, Chromosomes and Speciation (Cambridge Studies in Morphology and Molecules: New Paradigms in Evolutionary Bio); Searle, J., Polly, P., Zima, J., Eds.; Cambridge University Press: Cambridge, UK, 2019. [Google Scholar] [CrossRef]
- Piálek, J.; Hauffe, H.C.; Searle, J.B. Chromosomal variation in the house mouse. Biol. J. Linn. Soc. 2005, 84, 535–563. [Google Scholar] [CrossRef] [Green Version]
- White, T.A.; Bordewich, M.; Searl, J.B. A network approach to study karyotypic evolution: The chromosomal races of the common shrew (Sorex araneus) and house mouse (Mus musculus) as model systems. Syst. Biol. 2010, 59, 262–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulatova, N.Sh.; Pavlova, S.V.; Romanenko, S.A.; Serdyukova, N.A.; Golenishchev, F.N.; Malygin, V.M.; Lavrenchenko, L.A. Molecular cytogenetic markers of cryptic species and hybrids of the common Vole superspecies complex Microtus arvalis. Tsitologia 2013, 55, 268–271. [Google Scholar]
- Navarro, A.; Barton, N.H. Chromosomal speciation and molecular divergence—Accelerated evolution in rearranged chromosomes. Science 2003, 300, 321–324. [Google Scholar] [CrossRef] [Green Version]
- Mills, P.J.; Cook, L.G. Rapid chromosomal evolution in a morphologically cryptic radiation. Mol. Phylogenet. Evol. 2014, 77, 126–135. [Google Scholar] [CrossRef]
- Soldatović, B. Karyotype analysis and cytogenetic aspects of speciation in the genus Spalax. Zb. Prir. Nauk. Mat. Srp. 1977, 52, 5–58, (In Serbian with English Summary). [Google Scholar]
- Bugarski-Stanojević, V.; Stamenković, G.; Ćirović, D.; Ćirić, D.; Stojković, O.; Veličković, J.; Kataranovski, D.; Savić, I.R. 16S rRNA gene polymorphism supports cryptic speciation within the lesser blind mole rat Nannospalax leucodon superspecies (Rodentia: Spalacidae). Mamm. Biol. 2020, 100, 315–324. [Google Scholar] [CrossRef]
- Németh, A.; Csorba, G.; Laczkó, L.; Mizsei, E.; Bereczki, J.; Pásztor, J.A.; Petró, P.; Sramkó, G. Multi-locus genetic identification of a newly discovered population reveals a deep genetic divergence in European blind mole rats (Rodentia: Spalacidae: Nannospalax). Ann. Zool. Fenn. 2020, 57, 89–98. [Google Scholar] [CrossRef]
- Kryštufek, B.; Ivanitskaya, E.; Arslan, A.; Arslan, E.; Buzan, E. Evolutionary history of mole rats (genus Nannospalax) inferred from mitochondrial cytochrome b sequences. Biol. J. Linn. Soc. 2012, 105, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Hadid, Y.; Németh, A.; Snir, S.; Pavlíček, T.; Csorba, G.; Kázmér, M.; Major, A.; Mezhzherin, S.; Rusin, M.; Coşkun, Y.; et al. Is evolution of blind mole rats determined by climate oscillations? PLoS ONE 2012, 7, e30043. [Google Scholar] [CrossRef]
- Guha, G.; Goyal, S.P.; Kashyap, V.K. Molecular phylogeny of musk deer: A genomic view with mitochondrial 16S rRNA and cytochrome b gene. Mol. Phylogenet. Evol. 2007, 42, 585–597. [Google Scholar] [CrossRef]
- Caetano-Anollés, G. Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res. 2002, 30, 2575–2587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P., Jr. How Close Is Close: 16s RNA Sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992, 42, 166–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.; She, Y.; Elzo, M.A.; Zhang, C.; Fang, X.; Chen, H. Exploring genetic diversity and phylogenic relationships of Chinese cattle using gene mtDNA 16S rRNA. Arch. Anim. Breed. 2019, 62, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Németh, A.; Krnács, G.; Krizsik, V.; Révay, T.; Czabán, D.; Stojnić, N.; Farkas, J.; Csorba, G. European rodent on the edge: Status and distribution of the Vojvodina blind mole rat. Springerplus 2013, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chişamera, G.; Bužan, E.V.; Sahlean, T.; Murariu, D.; Zupan, S.; Krystufek, B. Bukovina blind mole rat Spalax graecus revisited: Phylogenetics, morphology, taxonomy, habitat associations and conservation. Mammal Rev. 2014, 44, 19–29. [Google Scholar] [CrossRef]
- Bugarski-Stanojević, V.; Jojić, V.; Stamenković, G.; Ćosić, N.; Ðokić, M.; Ćirović, D.; Savić, I.R. Cryptic subterranean mammal species, the lesser blind mole rat (Nannospalax leucodon syrmiensis)—Retreated but not extinct. Mamm. Biol. 2022; submitted. [Google Scholar]
- Veith, M.; Kosuch, J.; Vences, M. Climatic oscillations triggered post-Messinian speciation of Western Palearctic brown frogs (Amphibia, Ranidae). Mol. Phylogenet. Evol. 2003, 26, 310–327. [Google Scholar] [CrossRef]
- Bibb, M.J.; Van Etten, R.A.; Wright, C.T.; Walberg, M.W.; Clayton, D.A. Sequence and Gene Organization of Mouse Mitochondrial DNA. Cell 1981, 26, 167–180. [Google Scholar] [CrossRef]
- Schlegel, M.; Ali, H.S.; Stieger, N.; Groschup, M.H.; Wolf, R.; Ulrich, R.G. Molecular identification of small mammal species using novel cytochrome B gene-derived degenerated primers. Biochem. Genet. 2012, 50, 440–447. [Google Scholar] [CrossRef]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the Cytochrome b Gene of Mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Musser, G.G.; Carleton, M.D. Order Rodentia. In Mammal Species of the World: A Taxonomic and Geographic Reference; Wilson, D.E., Reeder, D.M., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 745–1601. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA Sequence Polymorphism Anaysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating Divergence Times in Large Molecular Phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Qiqing, T.; Kumar, S. Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Tao, Q.; Tamura, K.; Mello, B.; Kumar, S. Reliable Confidence Intervals for RelTime Estimates of Evolutionary Divergence Times. Mol. Biol. Evol. 2020, 37, 280–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savić, I.; Soldatović, B. Contribution to the study of ecogeographic distribution and evolution of chromosomal forms of the Spalacidae from the Balkan Peninsula. Arch. Biol. Sci. 1977, 29, 141–156, (In Serbian with English Summary). [Google Scholar]
- Akkiraz, M.S.; Akgün, F.; Utescher, T.; Bruch, A.A.; Mosbrugger, V. Precipitation gradients during the Miocene in Western and Central Turkey as quantified from pollen data. Palaeogeogr. Palaeocl. 2011, 304, 276–290. [Google Scholar] [CrossRef]
- Topachevski, V.A. The Fauna of the USSR: Mammals, Mole Rats, Spalacidae; Nauka: Leningrad, Russia, 1969; Volume 3, p. 99. (In Russian) [Google Scholar]
- De Bruijn, H. Remains of the mole rat Microspalax odessanus Topachevski from Karaburun (Greece, Macedonia) and the family Spalacidae. Proc. Koninklke Ned. Akad. Wet. Ser. B 1984, 87, 417–425. [Google Scholar]
- Ferguson-Smith, M.A.; Trifonov, V. Mammalian karyotype evolution. Nat. Rev. Genet. 2007, 8, 950–962. [Google Scholar] [CrossRef]
- Csorba, G.; Krivek, G.; Sendula, T.; Homonnay, Z.G.; Hegyeli, Z.; Sugár, S.; Farkas, J.; Stojnić, N.; Németh, A. How can scientific researches change conservation priorities? A review of decade-long research on blind mole-rats (Rodentia: Spalacinae) in the Carpathian Basin. Therya 2015, 6, 103–121. [Google Scholar] [CrossRef] [Green Version]
- Németh, A.; Révay, T.; Hegyeli, Z.; Farkas, J.; Czabán, D.; Rózsás, A.; Csorba, G. Chromosomal forms and risk assessment of Nannospalax (superspecies leucodon) (Mammalia: Rodentia) in the Carpathian Basin. Folia Zool. 2009, 58, 349–361. [Google Scholar]
- Savić, I.R.; Rempe, U. Vergleichende Kraniometrische Untersuchungen über Vertrerer der Gattung Spalax (Microspalax) auf der Balkan-Halbinsel; Hauptversammlung der Deutschen Gesellschaft für Säugetierkunde: Bamberg, Germany, 1977. [Google Scholar]
- Raicu, P.; Bratosin, S.; Hamar, M. Study on the karyotype of Spalax leucodon Nordm. and S. microphthalmus Güld. Caryologia 1968, 21, 127–135. [Google Scholar] [CrossRef]
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. USA 2005, 102, 6535–6542. [Google Scholar] [CrossRef] [Green Version]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Faria, R.; Neto, S.; Noor, M.A.F.; Navarro, A. Role of Natural Selection in Chromosomal Speciation; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar] [CrossRef]
- Colangelo, P.; Corti, M.; Verheyen, E.; Annesi, F.; Oguge, N.; Makundi, R.H.; Verheyen, W. Mitochondrial phylogeny reveals differential modes of chromosomal evolution in the genus Tatera (Rodentia: Gerbillinae) in Africa. Mol. Phylogenet. Evol. 2005, 35, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Pansonato-Alves, J.C.; Costa-Silva, G.J.; Mendonça, F.F.; Scacchetti, P.C.; Oliveira, C.; Foresti, F. Molecular and cytogenetic analyses of cryptic species within the Synbranchus marmoratus Bloch, 1795 (Synbranchiformes: Synbranchidae) grouping: Species delimitations, karyotypic evolution and intraspecific diversification. Neotrop. Ichthyol. 2014, 12, 903–911. [Google Scholar] [CrossRef] [Green Version]
- Baskevich, M.I.; Potapov, S.G.; Mironova, T.A. Caucasian Cryptic Species of Rodents as Models in Research on the Problems of Species and Speciation. Biol. Bull. Rev. 2016, 6, 245–259. [Google Scholar] [CrossRef]
- Malcher, S.M.; Pieczarka, J.C.; Geise, L.; Rossi, R.V.; Pereira, A.L.; O’Brien, P.C.M.; Asfora, P.H.; da Silva, V.F.; Sampaio, M.I.; Ferguson-Smith, M.A.; et al. Oecomys catherinae (Sigmodontinae, Cricetidae): Evidence for chromosomal speciation? PLoS ONE 2017, 12, e0181434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capanna, E.; Civitelli, M.V.; Cristaldi, M. Chromosomal rearrangement, reproductive isolation and speciation in mammals. The case of Mus musculus. Boll. Zool. 1977, 44, 213–246. [Google Scholar] [CrossRef]
- Rieseberg, L.H. Chromosomal rearrangements and Speciation. Trends Ecol. Evol. 2001, 16, 7. [Google Scholar] [CrossRef]
- Sözen, M.; Matur, F.; Çolak, E.; Özkurt, Ö.; Karataş, A. Some karyological records and a new chromosomal form for Spalax (Mammalia: Rodentia) in Turkey. Folia Zool. 2006, 55, 247–256. [Google Scholar]
- Soldatović, B.; Savić, I.R. New karyotype form of the mole rat (Spalax Güld.). Arch. Biol. Sci. 1973, 25, 13–14. [Google Scholar]
- Kandemir, I.; Sözen, M.; Matur, F.; Kankılıç, T.; Martínková, N.; Çolak, F.; Özkurt, S.Ö.; Çolak, E. Phylogeny of species and cytotypes of mole rats (Spalacidae) in Turkey inferred from mitochondrial cytochrome b sequences. Folia Zool. 2012, 61, 25–33. [Google Scholar] [CrossRef]
- Sözen, M.; Çolak, F.; Sevindik, M.; Matur, F. Cytotypes of Nannospalax xanthodon (Satunin, 1898) (Rodentia, Spalacidae) from western Anatolia. Turk. J. Zool. 2013, 37, 462–469. [Google Scholar] [CrossRef]
- Agapow, P.-M.; Bininda-Emonds, O.R.P.; Crandall, K.A.; Gittleman, J.L.; Mace, G.M.; Marshall, J.C.; Purvis, A. The impact of species concept on biodiversity studies. Q. Rev. Biol. 2004, 79, 161–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savić, I.; Nevo, E. The Spalacidae: Evolutionary history, speciation, and population biology. In Evolution of Subterranean Mammals at the Organismal and Molecular Levels; Nevo, E., Reig, O.A., Eds.; Alan R. Liss, Inc.: New York, NY, USA, 1990; pp. 129–153. [Google Scholar]
- Vasić, V.; Džukić, G.; Janković, D.; Simonov, N.; Petrov, B.; Savić, I. Preliminarni spisak vrsta za crvenu listu kičmenjaka Srbije. Zaštita Prir.—Prot. Nat. 1991, 43–44, 121–132, (In Serbian and Latin). [Google Scholar]
- IUCN. Threats Classification Scheme. Version 3.2. 2012. Available online: www.iucnredlist.org/resources/threat-classification-scheme (accessed on 12 November 2021).
- IUCN. Conservation Actions Classification Scheme. Version 2.0. 2012. Available online: www.iucnredlist.org/resources/conservation-actions-classification-scheme (accessed on 12 November 2021).
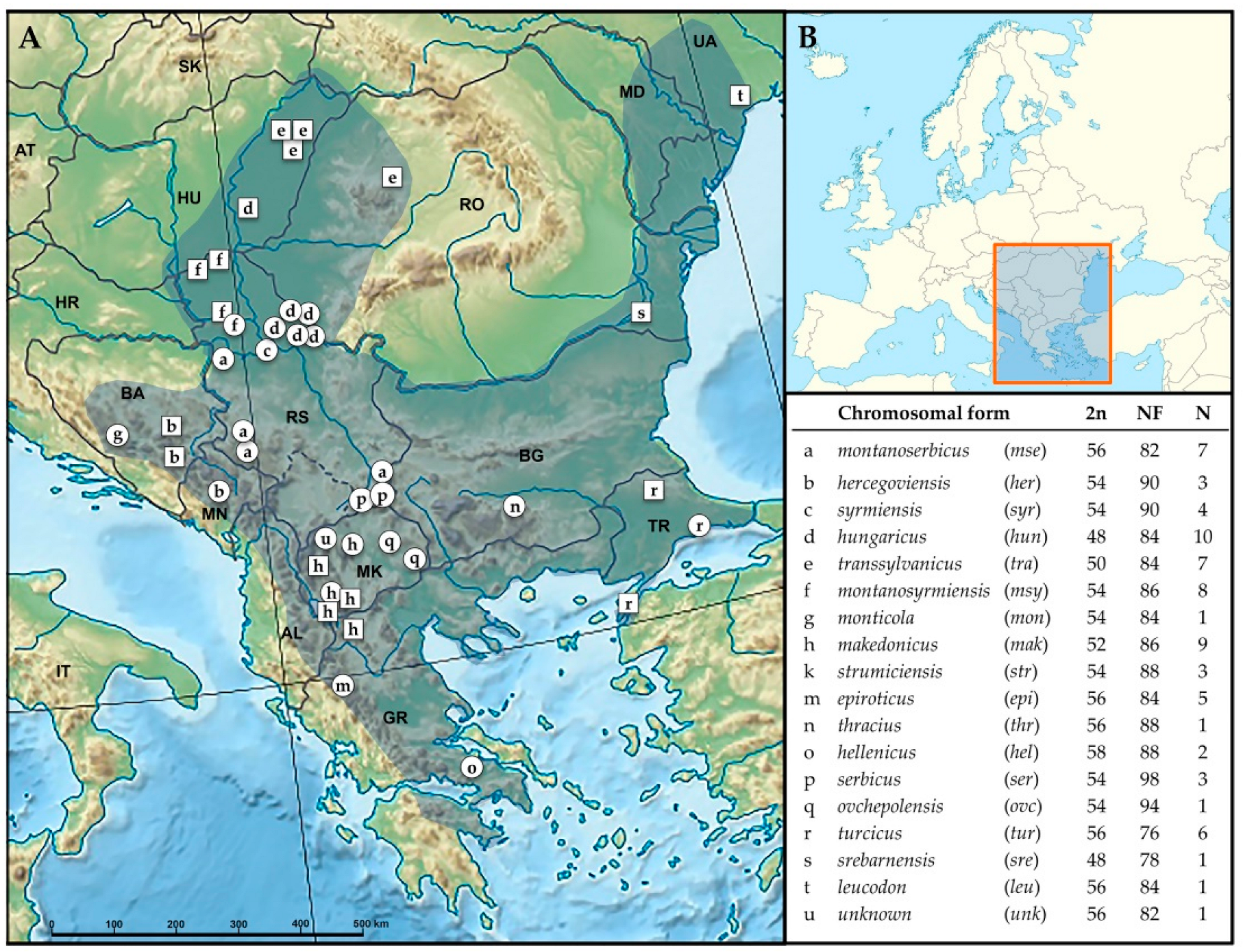
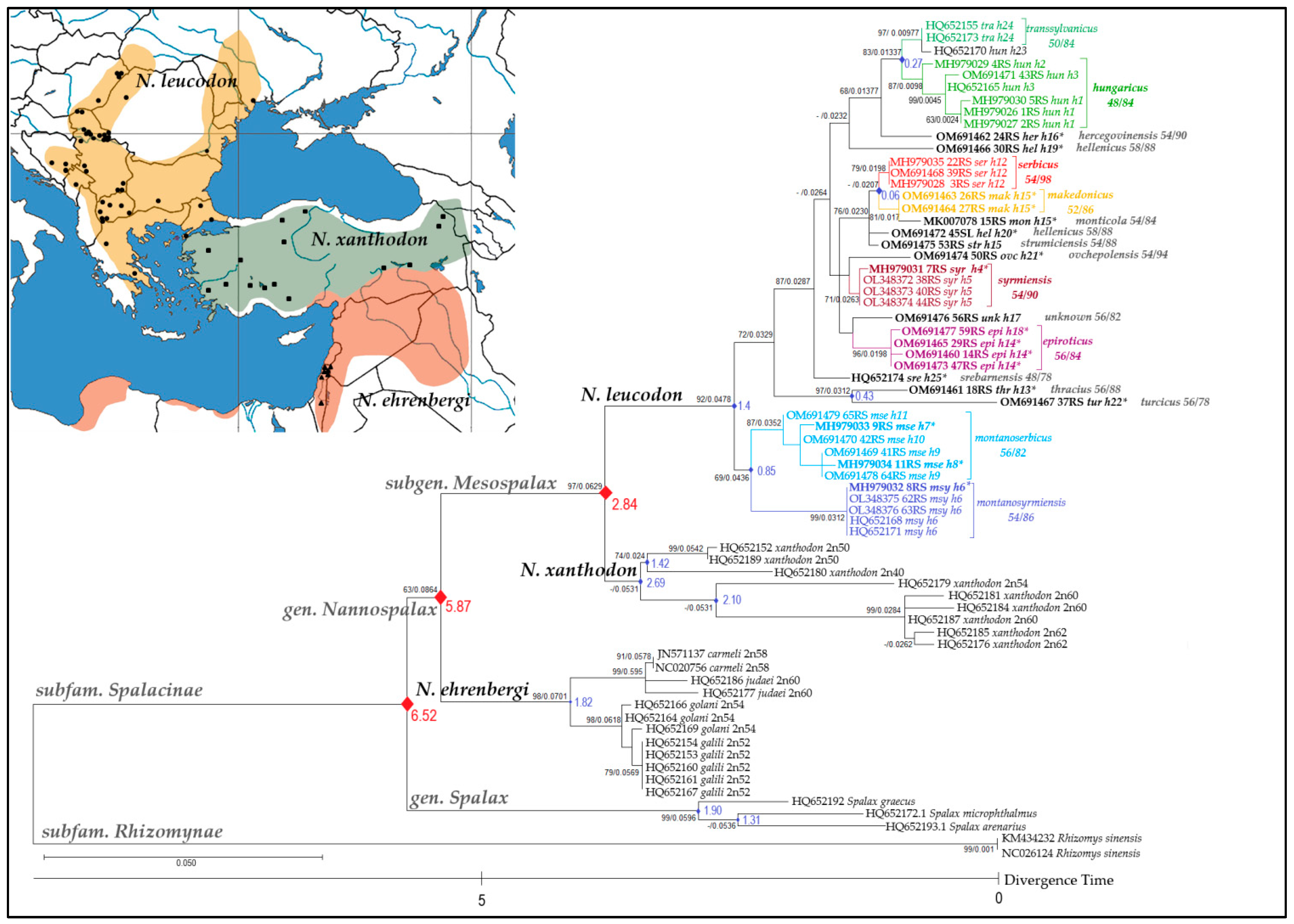
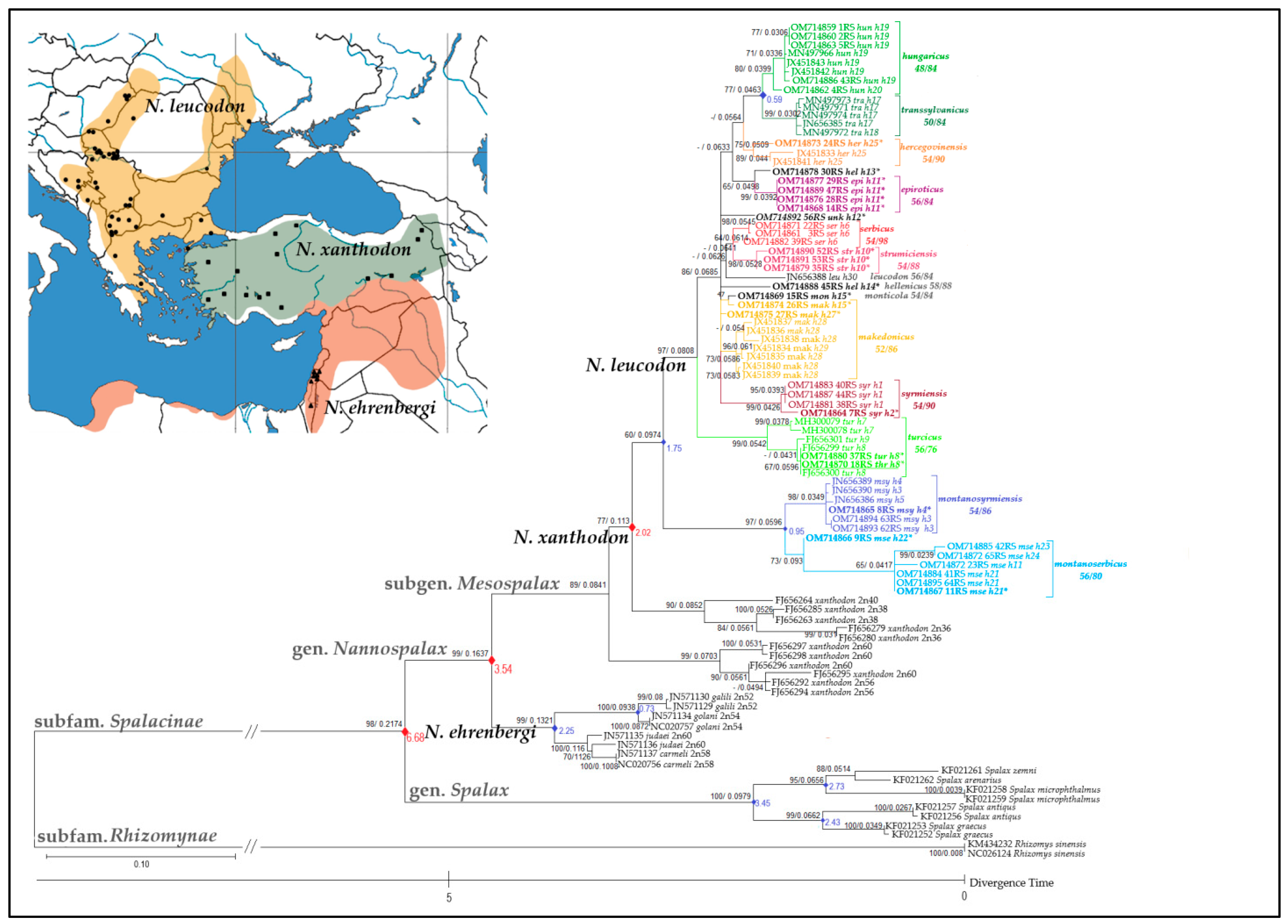
| 16S rRNA ID | MT-CYTB ID | Sample No | Sampling Locality b | Chromosomal Form | 2n/NF | Year of Capture | Tissue |
|---|---|---|---|---|---|---|---|
| MH979033 h 7 | OM714866 h 22 | 9RS a | Zlatibor Mt, RS | mse (a) | 56/82 | 1965 | Tooth |
| MH979034 h 8 | OM714867 h 21 | 11RS a | Vlasina, RS | mse (a) | 56/82 | 1965 | Tooth |
| / | OM714872 h 11 | 23RS | Vlasina, RS | mse (a) | 56/82 | 2018 | Muscle |
| OM691469 h 9 | OM714884 h 21 | 41RS | Vlasina, RS | mse (a) | 56/82 | 2020 | Liver |
| OM691470 h 10 | OM714885 h 23 | 42RS | Jadovnik Mt, RS | mse (a) | 56/82 | 2020 | Liver |
| OM691478 h 9 | OM714895 h 21 | 64RS | Vlasina, RS | mse (a) | 56/82 | 2021 | Liver |
| OM691479 h 11 | OM714896 h 24 | 65RS | Mačvanski Pričinović, RS | mse (a) | 56/82 | 2017 | Liver |
| OM691462 h 16 | OM714873 h 25 | 24RS a | Gvozd Mt, MN | her (b) | 54/90 | 1967 | Tooth |
| MH979031 h 4 | OM714864 h 2 | 7RS a | Beograd, RS | syr (c) | 54/90 | 1962 | Tooth |
| OL348372 h 5 | OM714881 h 1 | 38SL | Beograd, RS | syr (c) | 54/90 | 2019 | Liver |
| OL348373 h 5 | OM714883 h 1 | 40RS | Beograd, RS | syr (c) | 54/90 | 2020 | Liver |
| OL348374 h 5 | OM714887 h 1 | 44RS | Beograd, RS | syr (c) | 54/90 | 2021 | Liver |
| MH979026 h 1 | OM714859 h 19 | 1RS | Šumarak, RS | hun (d) | 48/84 | 2016 | Liver |
| MH979027 h 1 | OM714860 h 19 | 2RS | Šumarak, RS | hun (d) | 48/84 | 2016 | Liver |
| MH979029 h 2 | OM714862 h 20 | 4RS | Pančevo, RS | hun (d) | 48/84 | 2017 | Liver |
| MH979030 h 1 | OM714863 h 19 | 5RS | Kajtasovo, RS | hun (d) | 48/84 | 2018 | Liver |
| OM691471 h 3 | OM714886 h 19 | 43RS | Vršački Breg, RS | hun (d) | 48/84 | 2020 | Liver |
| MH979032 h 6 | OM714865 h 4 | 8RS a | Stražilovo, RS | msy (f) | 54/86 | 1965 | Tooth |
| OL348375 h 6 | OM714893 h 3 | 62RS | Sremski Karlovci, RS | msy (f) | 54/86 | 2021 | Liver |
| OL348376 h 6 | OM714894 h 3 | 63RS | Sremski Karlovci, RS | msy (f) | 54/86 | 2021 | Liver |
| MK007078 h 15 | OM714869 h 15 | 15RS a | Šuica, BA | mon (g) | 54/84 | 1979 | Tooth |
| OM691463 h 15 | OM714874 h 15 | 26RS a | Jakupica, MK | mak (h) | 52/86 | 1972 | Tooth |
| OM691464 h 15 | OM714875 h 27 | 27RS a | Ohrid, MK | mak (h) | 52/86 | 1975 | Tooth |
| / | OM714879 h 10 | 35RS a | Strumica, MK | str (k) | 54/88 | 1975 | Tooth |
| / | OM714890 h 10 | 52RS a | Strumica, MK | str (k) | 54/88 | 1975 | Tooth |
| OM691475 h 15 | OM714891 h 10 | 53RS a | Strumica, MK | str (k) | 54/88 | 1975 | Tooth |
| OM691460 h 14 | OM714868 h 11 | 14RS a | Lefkothea, GR | epi (m) | 56/84 | 1975 | Tooth |
| / | OM714876 h 11 | 28RS a | Lefkothea, GR | epi (m) | 56/84 | 1975 | Tooth |
| OM691465 h 14 | OM714877 h 11 | 29RS a | Lefkothea, GR | epi (m) | 56/84 | 1975 | Tooth |
| OM691473 h 14 | OM714889 h 11 | 47RS a | Lefkothea, GR | epi (m) | 56/84 | 1975 | Tooth |
| OM691477 h 18 | / | 59RS a | Lefkothea, GR | epi (m) | 56/84 | 1975 | Tooth |
| OM691461 h 13 | OM714870 h 8 | 18RS a | Novo Selo, BG | thr (n) | 56/76–78 | 1976 | Tooth |
| OM691466 h 19 | OM714878 h 13 | 30RS a | Levadia, GR | hel (o) | 58/88 | 1975 | Tooth |
| OM691472 h 20 | OM714888 h 14 | 45RS a | Levadia, GR | hel (o) | 58/88 | 1975 | Tooth |
| MH979028 h 12 | OM714861 h 6 | 3RS | Ristovac, RS | ser (p) | 54/98 | 2014 | Muscle |
| MH979035 h 12 | OM714871 h 6 | 22RS | Klinovac, RS | ser (p) | 54/98 | 2018 | Muscle |
| OM691468 h 12 | OM714882 h 6 | 39RS | Ristovac, RS | ser (p) | 54/98 | 2019 | Muscle |
| OM691474 h 21 | / | 50RS a | Štip, MK | ovc (q) | 54/94 | 1986 | Tooth |
| OM691467 h 22 | OM714880 h 8 | 37RS a | Çorlu, TR | tur (r) | 56/76–78 | 1976 | Tooth |
| OM691476 h 17 | OM714892 h 12 | 56RS a | Popova Šapka, MK | unk (u) | 56/82 | 1972 | Tooth |
| hun | syr | msy | mse | ser | her | mak | mon | unk | epi | str | hel | ovc | thr | tur | tra | sre | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hun | 0.006 | 0.009 | 0.009 | 0.007 | 0.006 | 0.007 | 0.009 | 0.006 | 0.007 | 0.007 | 0.006 | 0.009 | 0.011 | 0.012 | 0.004 | 0.008 | |
| syr | 0.027 | 0.009 | 0.008 | 0.005 | 0.006 | 0.005 | 0.006 | 0.005 | 0.005 | 0.004 | 0.004 | 0.002 | 0.009 | 0.010 | 0.006 | 0.005 | |
| msy | 0.045 | 0.040 | 0.006 | 0.010 | 0.010 | 0.010 | 0.012 | 0.008 | 0.009 | 0.009 | 0.009 | 0.012 | 0.010 | 0.011 | 0.009 | 0.009 | |
| mse | 0.040 | 0.033 | 0.023 | 0.009 | 0.009 | 0.009 | 0.011 | 0.008 | 0.009 | 0.009 | 0.008 | 0.010 | 0.009 | 0.010 | 0.009 | 0.008 | |
| ser | 0.025 | 0.014 | 0.044 | 0.039 | 0.007 | 0.003 | 0.005 | 0.005 | 0.006 | 0.003 | 0.004 | 0.004 | 0.010 | 0.010 | 0.006 | 0.006 | |
| her | 0.022 | 0.023 | 0.044 | 0.037 | 0.027 | 0.008 | 0.010 | 0.006 | 0.007 | 0.007 | 0.006 | 0.009 | 0.011 | 0.012 | 0.006 | 0.006 | |
| mak | 0.030 | 0.016 | 0.046 | 0.041 | 0.005 | 0.029 | 0.003 | 0.005 | 0.006 | 0.004 | 0.004 | 0.005 | 0.010 | 0.010 | 0.007 | 0.006 | |
| mon | 0.038 | 0.020 | 0.059 | 0.048 | 0.010 | 0.038 | 0.004 | 0.007 | 0.008 | 0.005 | 0.006 | 0.005 | 0.009 | 0.012 | 0.009 | 0.008 | |
| unk | 0.025 | 0.014 | 0.036 | 0.028 | 0.014 | 0.021 | 0.016 | 0.023 | 0.005 | 0.005 | 0.005 | 0.005 | 0.010 | 0.011 | 0.006 | 0.005 | |
| epi | 0.029 | 0.015 | 0.041 | 0.038 | 0.020 | 0.025 | 0.021 | 0.027 | 0.014 | 0.006 | 0.005 | 0.006 | 0.010 | 0.011 | 0.007 | 0.006 | |
| str | 0.026 | 0.012 | 0.038 | 0.037 | 0.005 | 0.025 | 0.007 | 0.012 | 0.012 | 0.018 | 0.004 | 0.003 | 0.010 | 0.011 | 0.007 | 0.006 | |
| hel | 0.029 | 0.018 | 0.044 | 0.039 | 0.017 | 0.026 | 0.017 | 0.024 | 0.020 | 0.020 | 0.015 | 0.004 | 0.009 | 0.010 | 0.006 | 0.005 | |
| ovc | 0.032 | 0.003 | 0.050 | 0.038 | 0.007 | 0.027 | 0.010 | 0.010 | 0.009 | 0.015 | 0.005 | 0.016 | 0.010 | 0.010 | 0.008 | 0.007 | |
| thr | 0.051 | 0.035 | 0.043 | 0.037 | 0.038 | 0.046 | 0.041 | 0.038 | 0.043 | 0.043 | 0.041 | 0.043 | 0.035 | 0.008 | 0.010 | 0.010 | |
| tur | 0.065 | 0.052 | 0.056 | 0.050 | 0.052 | 0.064 | 0.050 | 0.054 | 0.056 | 0.057 | 0.054 | 0.055 | 0.037 | 0.031 | 0.011 | 0.010 | |
| tra | 0.012 | 0.021 | 0.042 | 0.037 | 0.022 | 0.016 | 0.023 | 0.032 | 0.020 | 0.023 | 0.023 | 0.026 | 0.024 | 0.043 | 0.058 | 0.007 | |
| sre | 0.032 | 0.016 | 0.036 | 0.029 | 0.020 | 0.020 | 0.021 | 0.029 | 0.016 | 0.021 | 0.018 | 0.021 | 0.017 | 0.039 | 0.050 | 0.025 |
| N. l | N. x | N. e | S. g | S. m | S. a | R. s | |
|---|---|---|---|---|---|---|---|
| N. leucodon | 0.009 | 0.013 | 0.016 | 0.016 | 0.017 | 0.022 | |
| N. xanthodon | 0.066 | 0.012 | 0.016 | 0.015 | 0.016 | 0.021 | |
| N. ehrenbergi | 0.087 | 0.081 | 0.014 | 0.015 | 0.014 | 0.020 | |
| S. graecus | 0.107 | 0.113 | 0.087 | 0.009 | 0.010 | 0.023 | |
| S. microphthalmus | 0.110 | 0.112 | 0.099 | 0.041 | 0.010 | 0.022 | |
| S. arenarius | 0.115 | 0.118 | 0.094 | 0.045 | 0.045 | 0.022 | |
| R. sinensis | 0.173 | 0.172 | 0.153 | 0.172 | 0.164 | 0.167 |
| mse | mak | tur | tra | hel | syr | her | msy | epi | unk | ser | hun | mon | str | leu | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mse | 0.015 | 0.015 | 0.014 | 0.017 | 0.016 | 0.014 | 0.013 | 0.018 | 0.017 | 0.015 | 0.013 | 0.013 | 0.018 | 0.015 | |
| mak | 0.099 | 0.010 | 0.007 | 0.006 | 0.009 | 0.006 | 0.014 | 0.008 | 0.008 | 0.005 | 0.006 | 0.004 | 0.007 | 0.008 | |
| tur | 0.090 | 0.050 | 0.011 | 0.011 | 0.013 | 0.011 | 0.016 | 0.011 | 0.013 | 0.010 | 0.011 | 0.010 | 0.013 | 0.012 | |
| tra | 0.102 | 0.043 | 0.063 | 0.008 | 0.010 | 0.007 | 0.015 | 0.009 | 0.011 | 0.008 | 0.006 | 0.008 | 0.009 | 0.009 | |
| hel | 0.100 | 0.027 | 0.052 | 0.035 | 0.011 | 0.007 | 0.018 | 0.007 | 0.008 | 0.006 | 0.009 | 0.007 | 0.009 | 0.010 | |
| syr | 0.104 | 0.042 | 0.072 | 0.062 | 0.050 | 0.010 | 0.017 | 0.013 | 0.013 | 0.008 | 0.010 | 0.008 | 0.011 | 0.011 | |
| her | 0.088 | 0.031 | 0.062 | 0.037 | 0.033 | 0.052 | 0.016 | 0.009 | 0.008 | 0.006 | 0.006 | 0.005 | 0.009 | 0.008 | |
| msy | 0.071 | 0.093 | 0.094 | 0.103 | 0.100 | 0.108 | 0.102 | 0.018 | 0.018 | 0.016 | 0.014 | 0.014 | 0.019 | 0.017 | |
| epi | 0.102 | 0.028 | 0.049 | 0.031 | 0.025 | 0.055 | 0.033 | 0.092 | 0.009 | 0.009 | 0.010 | 0.008 | 0.011 | 0.012 | |
| unk | 0.080 | 0.020 | 0.051 | 0.035 | 0.030 | 0.048 | 0.027 | 0.081 | 0.024 | 0.009 | 0.010 | 0.007 | 0.011 | 0.012 | |
| ser | 0.097 | 0.022 | 0.050 | 0.044 | 0.024 | 0.038 | 0.033 | 0.099 | 0.031 | 0.025 | 0.007 | 0.005 | 0.006 | 0.007 | |
| hun | 0.093 | 0.033 | 0.062 | 0.029 | 0.040 | 0.053 | 0.032 | 0.092 | 0.038 | 0.033 | 0.038 | 0.006 | 0.010 | 0.008 | |
| mon | 0.091 | 0.017 | 0.049 | 0.041 | 0.026 | 0.041 | 0.026 | 0.097 | 0.027 | 0.018 | 0.019 | 0.029 | 0.008 | 0.007 | |
| str | 0.108 | 0.029 | 0.063 | 0.045 | 0.036 | 0.053 | 0.040 | 0.111 | 0.045 | 0.033 | 0.018 | 0.047 | 0.028 | 0.009 | |
| leu | 0.108 | 0.043 | 0.071 | 0.056 | 0.046 | 0.061 | 0.044 | 0.118 | 0.046 | 0.043 | 0.037 | 0.049 | 0.037 | 0.041 |
| N. e | N. l | S. z | N. x | S. a | R. s | S. ar | S. m | S. g | |
|---|---|---|---|---|---|---|---|---|---|
| N. ehrenbergi | 0.012 | 0.017 | 0.015 | 0.016 | 0.021 | 0.017 | 0.018 | 0.017 | |
| N. leucodon | 0.129 | 0.017 | 0.012 | 0.018 | 0.023 | 0.018 | 0.018 | 0.018 | |
| S. zemni | 0.201 | 0.194 | 0.021 | 0.014 | 0.022 | 0.009 | 0.012 | 0.013 | |
| N. xanthodon | 0.123 | 0.100 | 0.188 | 0.022 | 0.029 | 0.022 | 0.022 | 0.021 | |
| S. antiqus | 0.187 | 0.199 | 0.120 | 0.196 | 0.022 | 0.013 | 0.014 | 0.010 | |
| R. synensis | 0.266 | 0.287 | 0.286 | 0.292 | 0.274 | 0.022 | 0.023 | 0.022 | |
| S. arenarius | 0.200 | 0.205 | 0.055 | 0.193 | 0.110 | 0.282 | 0.011 | 0.012 | |
| S. microphthalmus | 0.218 | 0.211 | 0.101 | 0.192 | 0.131 | 0.306 | 0.084 | 0.014 | |
| S. graecus | 0.196 | 0.205 | 0.112 | 0.185 | 0.067 | 0.290 | 0.094 | 0.126 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bugarski-Stanojević, V.; Stamenković, G.; Jojić, V.; Ćosić, N.; Ćirović, D.; Stojković, O.; Veličković, J.; Savić, I. Cryptic Diversity of the European Blind Mole Rat Nannospalax leucodon Species Complex: Implications for Conservation. Animals 2022, 12, 1097. https://doi.org/10.3390/ani12091097
Bugarski-Stanojević V, Stamenković G, Jojić V, Ćosić N, Ćirović D, Stojković O, Veličković J, Savić I. Cryptic Diversity of the European Blind Mole Rat Nannospalax leucodon Species Complex: Implications for Conservation. Animals. 2022; 12(9):1097. https://doi.org/10.3390/ani12091097
Chicago/Turabian StyleBugarski-Stanojević, Vanja, Gorana Stamenković, Vida Jojić, Nada Ćosić, Duško Ćirović, Oliver Stojković, Jelena Veličković, and Ivo Savić. 2022. "Cryptic Diversity of the European Blind Mole Rat Nannospalax leucodon Species Complex: Implications for Conservation" Animals 12, no. 9: 1097. https://doi.org/10.3390/ani12091097
APA StyleBugarski-Stanojević, V., Stamenković, G., Jojić, V., Ćosić, N., Ćirović, D., Stojković, O., Veličković, J., & Savić, I. (2022). Cryptic Diversity of the European Blind Mole Rat Nannospalax leucodon Species Complex: Implications for Conservation. Animals, 12(9), 1097. https://doi.org/10.3390/ani12091097






