Study of the Fatty Acid Profile of Milk in Different Sheep Breeds: Evaluation by Multivariate Factorial Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Breeds and Dairy Systems
2.2. Fatty Acid Composition
2.3. Statistical Analysis
3. Results and Discussion
3.1. Milk FA Composition
3.2. Multivariate Factor Analysis
3.3. Effect of Breed on the Extracted Factor Scores
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Signorelli, F.; Contarini, G.; Annicchiarico, G.; Napolitano, F.; Orrù, L.; Catillo, G.; Haenlein, G.F.W.; Moioli, B. Breed differences in sheep milk fatty acid profiles: Opportunities for sustainable use of animal genetic resources. Small Rumin. Res. 2008, 78, 24–31. [Google Scholar] [CrossRef]
- Sanz Sampelayo, M.R.; Chilliard, Y.; Schmidely, P.; Boza, J. Influence of type of diet on the fat constituents of goat and sheep milk. Small Rumin. Res. 2007, 68, 42–63. [Google Scholar] [CrossRef]
- Nudda, A.; Mele, M.; Battacone, G.; Usai, M.G.; Macciotta, N.P.P. Comparison of conjugated linoleic acid (CLA) content in milk of ewes and goats with the same dietary regimen. Ital. J. Anim. Sci. 2003, 2, 515–517. [Google Scholar]
- Pulina, G.; Milán, M.J.; Lavín, M.P.; Theodoridis, A.; Morin, E.; Capote, J.; Thomas, D.L.; Francesconi, A.H.D.; Caja, G. Invited review: Current production trends, farm structures, and economics of the dairy sheep and goat sectors. J. Dairy Sci. 2018, 101, 6715–6729. [Google Scholar] [CrossRef] [PubMed]
- Carta, A.; Casu, S.; Usai, M.G.; Addis, M.; Fiori, M.; Fraghì, A.; Miari, S.; Mura, L.; Piredda, G.; Schibler, L.; et al. Investigating the genetic component of fatty acid content in sheep milk. Small Rumin. Res. 2008, 79, 22–28. [Google Scholar] [CrossRef]
- Macciotta, N.P.P.; Mele, M.; Cappio-Borlino, A.; Secchiari, P. Issues and perspectives in dairy sheep breeding. Ital. J. Anim. Sci. 2005, 4, 5–23. [Google Scholar] [CrossRef][Green Version]
- Astruc, J.M.; Barillet, F.; Carta, A.; Fioretti, M.; Gootwine, E.; Kompan, D.; Romberg, F.J.; Ugarte, E. Identification, Breeding, Production, Health and Recording on Farm Animals. In Proceedings of the 36th Biennial Session of the International Committee for Animal Recording (ICAR), Niagara Falls, NY, USA, 16–20 June 2008; Report of the Working Group on Milk Recording of Sheep, ICAR Technical Series No. 13. Slatter, J.D., Ed.; ICAR: Rome, Italy, 2008; pp. 275–282. [Google Scholar]
- Carta, A.; Casu, S.; Salaris, S. Invited review: Current state of genetic improvement in dairy sheep. J. Dairy Sci. 2009, 92, 5814–5833. [Google Scholar] [CrossRef] [PubMed]
- Rupp, R.; Mucha, S.; Larroque, H.; McEwan, J.; Conington, J. Genomic application in sheep and goat breeding. Anim. Front. 2016, 6, 39–44. [Google Scholar] [CrossRef]
- Cabiddu, A.; Decandia, M.; Addis, M.; Piredda, G.; Pirisi, A.; Molle, G. Managing Mediterranean pastures in order to enhance the level of beneficial fatty acids in sheep milk. Small Rumin. Res. 2005, 59, 169–180. [Google Scholar] [CrossRef]
- Sánchez, J.P.; San Primitivo, F.; Barbosa, E.; Varona, L.; De La Fuente, L.F. Genetic determination of fatty acid composition in Spanish Churra sheep milk. J. Dairy Sci. 2010, 93, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Stoop, W.M.; van Arendonk, J.A.M.; Heck, J.M.L.; van Valenberg, H.J.F.; Bovenhuis, G. Genetic parameters for major milk fatty acids and milk production traits of Dutch Holstein- Friesians. J. Dairy Sci. 2008, 91, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Pegolo, S.; Cecchinato, A.; Casellas, J.; Conte, G.; Mele, M.; Schiavon, S.; Bittante, G. Genetic and environmental relationships of detailed milk fatty acids profile determined by gas chromatography in Brown Swiss cows. J. Dairy Sci. 2016, 99, 1315–1330. [Google Scholar] [CrossRef] [PubMed]
- Correddu, F.; Cellesi, M.; Serdino, J.; Manca, M.G.; Contu, M.; Dimauro, C.; Ibba, I.; Macciotta, N.P.P. Genetic parameters of milk fatty acid profile in sheep: Comparison between gas chromatographic measurements and Fourier-Transform Infrared Spectroscopy predictions. Animal 2019, 13, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Stoop, W.M.; Bovenhuis, H.; Heck, J.M.L.; van Arendonk, J.A. Effect of lactation stage and energy status on milk fat composition of Holstein-Friesian cows. J. Dairy Sci. 2009, 92, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Bovenhuis, H.; Visker, M.H.; van Arendonk, J.A. Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, B.; Janss, L.L.; Poulsen, N.A.; Larsen, L.B.; Larsen, N.K.; Sørensen, P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genom. 2014, 15, 1112. [Google Scholar] [CrossRef] [PubMed]
- Crisà, A.; Marchitelli, C.; Pariset, L.; Contarini, G.; Signorelli, F.; Napolitano, F.; Catillo, G.; Valentini, A.; Moioli, B. Exploring polymorphisms and effects of candidate genes on milk fat quality in dairy sheep. J. Dairy Sci. 2010, 93, 3834–3845. [Google Scholar] [CrossRef] [PubMed]
- Moioli, B.; Contarini, G.; Pariset, L.; Marchitelli, C.; Crisà, A.; Catillo, G.; Napolitano, F. Genetic variation of C18:1 and C18:2 isomers in sheep milk fat. Small Rumin. Res. 2012, 103, 187–193. [Google Scholar] [CrossRef]
- Cesarani, A.; Sechi, T.; Gaspa, G.; Usai, M.G.; Sorbolini, S.; Macciotta, N.P.P.; Carta, A. Investigation of genetic diversity and selection signatures between Sarda T and Sardinian Ancestral black, two related sheep breeds with evident morphological differences. Small Rumin. Res. 2019, 177, 68–75. [Google Scholar] [CrossRef]
- Conte, G.; Serra, A.; Cremonesi, P.; Chessa, S.; Castiglioni, B.; Cappucci, A.; Bulleri, E.; Mele, M. Investigating mutual relationship among milk fatty acids by multivariate factor analysis in dairy cows. Livest. Sci. 2016, 188, 124–132. [Google Scholar] [CrossRef]
- Mele, M.; Macciotta, N.P.P.; Cecchinato, A.; Conte, G.; Schiavon, S.; Bittante, G. Multivariate factor analysis of detailed milk fatty acid profile: Effects of dairy system, feeding, herd, parity, and stage of lactation. J. Dairy Sci. 2016, 99, 9820–9833. [Google Scholar] [CrossRef] [PubMed]
- Palombo, V.; Conte, G.; Mele, M.; Macciotta, N.P.P.; Stefanon, B.; Ajmone Marsan, P.; D’Andrea, M. Use of multivariate factor analysis of detailed milk fatty acid profile to perform a genome-wide association study in Italian Simmental and Italian Holstein. J. Appl. Gen. 2020, 60, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Correddu, F.; Serdino, J.; Manca, M.G.; Cosenza, G.; Pauciullo, A.; Ramunno, L.; Macciotta, N.P.P. Use of multivariate factor analysis to characterize the fatty acid profile of buffalo milk. J. Food Compos. Anal. 2017, 60, 25–31. [Google Scholar] [CrossRef]
- Palombo, V.; Gaspa, G.; Conte, G.; Pilla, F.; Macciotta, N.P.P.; Mele, M.; D’Andrea, M. Combined multivariate factor analysis and GWAS for milk fatty acids trait in Comisana sheep breed. Anim. Genet. 2020, 51, 630–631. [Google Scholar] [CrossRef] [PubMed]
- Correddu, F.; Cesarani, A.; Dimauro, C.; Gaspa, G.; Macciotta, N.P.P. Principal component and multivariate factor analysis of detailed sheep milk fatty acid profile. J. Dairy Sci. 2021, 104, 5079–5094. [Google Scholar] [CrossRef] [PubMed]
- Mele, M.; Serra, A.; Buccioni, A.; Conte, G.; Pollicardo, A.; Secchiari, P. Effect of soybean oil supplementation on milk fatty acid composition from Saanen goats fed diets with different forage:concentrate ratios. Ital. J. Anim. Sci. 2008, 7, 297–311. [Google Scholar] [CrossRef]
- Kramer, J.K.G.; Cruz-Hernandez, C.; Deng, Z.Y.; Zhou, J.Q.; Jahreis, G.; Dugan, M.E.R. Analysis of conjugated linoleic acid and trans 18:1 isomers in synthetic and animal products. Am. J. Clin. Nutr. 2004, 79, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Morrison, F. Multivariate Statistical Methods; McGraw-Hill: New York, NY, USA, 1976. [Google Scholar]
- Kaiser, H.F.; Rice, J. Little jiffy, Mark IV. Educ. Psychol. Meas. 1974, 34, 111–117. [Google Scholar] [CrossRef]
- Macciotta, N.P.P.; Dimauro, C.; Null, D.J.; Gaspa, G.; Cellesi, M.; Cole, J.B. Dissection of genomic correlation matrices of US Holsteins using multivariate factor analysis. J. Anim. Breed Genet. 2015, 132, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Vallas, M.; Bovenhuis, H.; Kaart, T.; Pärna, K.; Kiiman, H.; Pärna, E. Genetic parameters for milk coagulation properties in Estonian Holstein cows. J. Dairy Sci. 2010, 93, 3789–3796. [Google Scholar] [CrossRef]
- Macciotta, N.P.P.; Cecchinato, A.; Mele, M.; Bittante, G. Use of multivariate factor analysis to define new indicator variables for milk composition and coagulation properties in Brown Swiss cows. J. Dairy Sci. 2012, 95, 7346–7354. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; Pryce, J.E.; Hayes, B.J.; Goddard, M.E. Multivariate analysis of genome-wide association study in cattle. J. Dairy Sci. 2010, 93, 3818–3833. [Google Scholar] [CrossRef] [PubMed]
- Cerny, B.A.; Kaiser, H.F. A study of a measure of sampling adequacy for factor analytic correlation matrices. Multivar. Behav. Res. 1977, 12, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Pontier, D.; Dufour, A.B. Genetic markers in the playground of multivariate analysis. Heredity 2009, 102, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Vlaeminck, B.; Fievez, V.; Cabrita, A.R.J.; Fonseca, A.J.M.; Dewhurst, R.J. Factors affecting odd- and branched-chain fatty acids in milk: A review. Anim. Feed Sci. Technol. 2006, 131, 389–417. [Google Scholar] [CrossRef]
- Cabiddu, A.; Peratoner, G.; Valenti, B.; Monteils, V.; Martin, B.; Coppa, M. A quantitative review of on-farm feeding practices to enhance the quality of grassland-based ruminant dairy and meat products. Animal 2021, 16, 100375. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Fievez, V.; Van Laar, H.; Demeyer, D. Rumen odd and branched chain fatty acids in relation to in vitro rumen volatile fatty acid productions and dietary characteristics of incubated substrates. J. Anim. Physiol. Anim. Nutr. 2004, 88, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Civico, A.; Sánchez, N.N.; Gómez-Cortés, P.; de la Fuente, M.A.; Blanco, F.P.; Juárez, M.; Schiavone, A.; Marin, A.L.M. Odd- and branched-chain fatty acids in goat milk as indicators of the diet composition. Ital. J. Anim. Sci. 2017, 16, 68–74. [Google Scholar] [CrossRef]
- Zhao, W.S.; Hu, S.L.; Yu, K.; Wang, H.; Wang, W.; Loor, J.J.; Luo, J. Lipoprotein Lipase, Tissue Expression and Effects on Genes Related to Fatty Acid Synthesis in Goat Mammary Epithelial Cells. Int. J. Mol. Sci. 2014, 15, 22757–22771. [Google Scholar] [CrossRef]
- Chilliard, Y.; Ferlay, A.; Mansbridge, R.M.; Doreau, M. Ruminant milk fat plasticity: Nutritional control of saturated, polyunsaturated, trans and conjugated fatty acids. Ann. Zootech. 2000, 49, 181–205. [Google Scholar] [CrossRef]
- Timmen, H.; Patton, S. Milk fat globules: Fatty acid composition, size and in vivo regulation of fat liquidity. Lipids 1988, 23, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Shingfield, K.J.; Bernard, L.; Leroux, C.; Chilliard, Y. Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal 2010, 4, 1140–1166. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, P.J. Nutritional Ecology of the Ruminant, 2nd ed.; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Leat, W.M.F.; Kemp, P.; Lysons, R.J.; Alexander, T.J.L. Fatty acid composition of depot fats from gnotobiotic lambs. J. Agric. Sci. 1977, 88, 175–179. [Google Scholar] [CrossRef]
- Griinari, J.M.; Dwyer, D.A.; McGuire, M.A.; Bauman, D.E.; Palmquist, D.L.; Nurmela, K.V.V. Trans-octadecenoic acids and milk fat depression in lactating dairy cows. J. Dairy Sci. 1998, 81, 1251–1261. [Google Scholar] [CrossRef]
- Griinari, J.M.; Bauman, D.E. Biosynthesis of conjugated linoleic acid and its incorporation into meat and milk in ruminants. In Advances in Conjugated Linoleic Acid Research; Yurawecz, M.P., Mossoba, M.M., Kramer, J.K.G., Pariza, M.W., Nelson, G.J., Eds.; AOCS Press: Champaign, IL, USA, 1999; Volume 1, pp. 180–200. [Google Scholar]
- Toral, P.G.; Hervás, G.; Frutos, P. Effect of lipid supplementation on the endogenous synthesis of milk cis-9,trans-11 conjugated linoleic acid in dairy sheep and goats: A tracer assay with 13C-vaccenic acid. J. Dairy Sci. 2022, 105, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Conte, G.; Serra, A.; Mele, M. Dairy cow breeding and feeding on the milk fatty acid pattern. In Nutrients in Dairy and Their Implications for Health and Disease; Watson, R.R., Collier, R.J., Preedy, V.R., Eds.; Academic Press: London, UK, 2017; pp. 19–41. [Google Scholar]
- Mele, M.; Conte, G.; Castiglioni, B.; Chessa, S.; Macciotta, N.P.P.; Serra, A.; Buccioni, A.; Pagnacco, G.; Secchiari, P. Stearoyl-CoA desaturase gene polymorphism and milk fatty acid composition in Italian Friesian cows. J. Dairy Sci. 2007, 90, 4458–4465. [Google Scholar] [CrossRef] [PubMed]
- Loften, J.R.; Linn, J.G.; Drackley, J.K.; Jenkins, T.C.; Soderholm, C.G.; Kertz, A.F. Invited review: Palmitic and stearic acid metabolism in lactating dairy cows. J. Dairy Sci. 2014, 97, 4661–4674. [Google Scholar] [CrossRef] [PubMed]
- Dann, H.M.; Morin, D.E.; Bollero, G.A.; Murphy, M.R.; Drackley, J.K. Prepartum intake, postpartum induction of ketosis, and periparturient disorders affect the metabolic status of dairy cows. J. Dairy Sci. 2005, 88, 3249–3264. [Google Scholar] [CrossRef]
- Kay, J.K.; Weber, W.J.; Moore, C.E.; Bauman, D.E.; Hansen, L.B.; Chester-Jones, H.; Crooker, B.A.; Baumgard, L.H. Effects of week of lactation and genetic selection for milk yield on milk fatty acid composition in Holstein cows. J. Dairy Sci. 2005, 88, 3886–3893. [Google Scholar] [CrossRef]
- Bionaz, M.; Chen, S.; Khan, M.J.; Loor, J.J. Functional role of PPARs in ruminants: Potential targets for fine-tuning metabolism during growth and lactation. PPAR Res. 2013, 1–28. [Google Scholar] [CrossRef]
- Turini, L.; Conte, G.; Bonelli, F.; Serra, A.; Sgorbini, M.; Mele, M. Multivariate factor analysis of milk fatty acid composition in relation to the somatic cell count of single udder quarters. J. Dairy Sci. 2020, 103, 7392–7406. [Google Scholar] [CrossRef] [PubMed]
- Marín, A.M.; Gómez-Cortés, P.; Castro, A.G.; Juárez, M.; Alba, L.P.; Hernández, M.P.; De la Fuente, M.A. Short communication: Linear discriminant analysis and type of oil added to dairy goat diets. J. Dairy Sci. 2012, 95, 4045–4049. [Google Scholar] [CrossRef] [PubMed]
- Dewhurst, R.J.; Shingfield, K.J.; Lee, M.R.F.; Scollan, N.D. Increasing the concentrations of beneficial polyunsaturated fatty acids in milk produced by dairy cows in high-forage systems. Anim. Feed Sci. Technol. 2006, 131, 168–206. [Google Scholar] [CrossRef]
- Nantapo, C.T.W.; Muchenje, V.; Hugo, A. Atherogenicity index and health-related fatty acids in different stages of lactation from Friesian, Jersey and Friesian × Jersey cross cow milk under a pasture-based dairy system. Food Chem. 2014, 146, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Roca Fernandez, A.I.; Gonzalez Rodriguez, A. Effect of Dietary and Animal Factors on Milk Fatty Acids Composition of Grazing Dairy Cows: A Review. Iran. J. Appl. Anim. Sci. 2012, 2, 97–109. [Google Scholar]
- Tsiplakou, E.; Kominakis, A.; Zervas, G. The interaction between breed and diet on CLA and fatty acid content of milk of four sheep breeds kept indoor or at grass. Small Rumin. Res. 2008, 74, 179–187. [Google Scholar] [CrossRef]
- Hanuš, O.; Samková, E.; Křížová, L.; Hasoňová, L.; Kala, R. Role of fatty acids in milk fat and the influence of selected factors on their variability—A review. Molecules 2018, 23, 1636. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, M.L.; Cersosimo, L.M.; Wright, A.D.G.; Kraft, J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 2016, 92, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.; Cabiddu, A.; Pinna, G.; Decandia, M.; Piredda, G.; Pirisi, A.; Molle, G. Milk and cheese fatty acid composition in sheep fed Mediterranean forages with reference to conjugated linoleic acid cis9,trans11. J. Dairy Sci. 2006, 88, 3443–3454. [Google Scholar] [CrossRef]
- Soyeurt, H.; Gillon, A.; Vanderick, S.; Mayeres, P.; Bertozzi, C.; Gengler, N. Estimation of heritability and genetic correlations for the major fatty acids in bovine milk. J. Dairy Sci. 2007, 90, 4435–4442. [Google Scholar] [CrossRef]
- Mele, M.; Dal Zotto, R.; Cassandro, M.; Conte, G.; Serra, A.; Buccioni, A.; Bittante, G.; Secchiari, P. Genetic parameters for conjugated linoleic acid, selected milk fatty acids, and milk fatty acid unsaturation of Italian Holstein-Friesian cows. J. Dairy Sci. 2009, 92, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Neto, L.R.P.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef] [PubMed]
- Ciani, E.; Crepaldi, P.; Nicoloso, L.; Lasagna, E.; Sarti, F.M.; Moioli, B.; Napolitano, F.; Carta, A.; Usai, G.; D’Andrea, M.; et al. Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim. Genet. 2013, 45, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Mele, M.; Contarini, G.; Cercaci, L.; Serra, A.; Buccioni, A.; Povolo, M.; Conte, G.; Funaro, A.; Banni, S.; Lercker, G.; et al. Enrichment of Pecorino cheese with conjugated linoleic acid by feeding dairy ewes with extruded linseed: Effect on fatty acid and triglycerides composition and on oxidative stability. Int. Dairy J. 2011, 21, 365–372. [Google Scholar] [CrossRef]
- Daghio, M.; Ciucci, F.; Buccioni, A.; Cappucci, A.; Casarosa, L.; Serra, A.; Conte, G.; Viti, C.; McAmmond, B.M.; Van Hamme, J.D.; et al. Correlation of breed, growth performance, and rumen microbiota in two rustic cattle breeds reared under different conditions. Front. Microbiol. 2021, 12, 652031. [Google Scholar] [CrossRef] [PubMed]
| Mean | SD | CV% | P5 | P95 | Kurtosis | |
|---|---|---|---|---|---|---|
| C4:0 | 3.24 | 0.81 | 25.03 | 2.32 | 4.90 | 0.95 |
| C6:0 | 1.91 | 0.41 | 21.41 | 1.23 | 2.56 | −0.14 |
| C8:0 | 1.78 | 0.44 | 24.71 | 1.11 | 2.54 | 0.24 |
| C10:0 | 5.52 | 1.63 | 29.54 | 3.05 | 8.45 | 0.36 |
| C10:1c9 | 0.20 | 0.09 | 43.61 | 0.06 | 0.34 | 0.45 |
| C12:0 | 3.28 | 0.91 | 27.69 | 2.07 | 4.91 | 0.91 |
| C13:0 | 0.06 | 0.03 | 51.68 | 0.02 | 0.10 | 1.97 |
| C14:0 | 9.70 | 1.79 | 18.51 | 7.25 | 12.78 | 0.43 |
| C14:0iso | 0.14 | 0.05 | 34.90 | 0.08 | 0.23 | 0.51 |
| C14:1c9 | 0.24 | 0.15 | 63.27 | 0.09 | 0.61 | 1.58 |
| C15:0 | 1.20 | 0.28 | 23.27 | 0.69 | 1.61 | −0.21 |
| C16:0iso | 0.33 | 0.10 | 30.02 | 0.17 | 0.49 | 0.22 |
| C16:0 | 23.11 | 3.42 | 14.78 | 17.52 | 28.92 | 0.05 |
| C16:1c9 | 0.74 | 0.26 | 35.63 | 0.35 | 1.20 | 0.88 |
| C18:0 | 9.82 | 2.08 | 21.13 | 6.77 | 13.24 | 0.81 |
| C18:1t6–8 | 0.24 | 0.13 | 54.51 | 0.10 | 0.55 | 2.17 |
| C18:1t9 | 0.28 | 0.11 | 39.36 | 0.16 | 0.52 | 2.04 |
| C18:1t10 | 0.41 | 0.19 | 45.82 | 0.19 | 0.75 | 2.24 |
| C18:1t11 | 2.33 | 1.45 | 62.08 | 0.83 | 4.74 | 2.89 |
| C18:1c9 | 18.25 | 3.43 | 18.81 | 12.75 | 23.60 | 0.01 |
| C18:1t15 | 0.43 | 0.16 | 36.70 | 0.17 | 0.70 | 0.16 |
| C18:1c12 | 0.27 | 0.10 | 37.23 | 0.11 | 0.40 | 1.50 |
| C18:2n6 | 2.18 | 0.55 | 24.99 | 1.30 | 3.05 | 0.32 |
| C20:0 | 0.28 | 0.09 | 31.94 | 0.15 | 0.43 | 0.45 |
| C18:3n3 | 1.10 | 0.49 | 44.54 | 0.39 | 2.09 | 0.96 |
| C18:2c9t11 | 1.28 | 0.58 | 45.63 | 0.50 | 2.26 | 1.37 |
| C20:1c11 | 0.04 | 0.02 | 57.62 | 0.01 | 0.08 | 1.90 |
| C21:0 | 0.09 | 0.04 | 40.67 | 0.02 | 0.14 | 0.01 |
| C20:4 n6 | 0.14 | 0.06 | 46.03 | 0.06 | 0.25 | 0.55 |
| C20:5 n3 | 0.07 | 0.02 | 36.37 | 0.03 | 0.11 | 0.53 |
| C24:0 | 0.06 | 0.03 | 42.39 | 0.03 | 0.11 | 0.95 |
| C22:5 n3 | 0.14 | 0.05 | 37.85 | 0.07 | 0.23 | 0.47 |
| C22:6 n3 | 0.06 | 0.04 | 66.26 | 0.00 | 0.14 | 0.91 |
| Name of Factors | OBCFA and LCFA | sn3_ Position | Alternative BH | SCD_1 | SCD_2 | SCD_3 | Fat Secretion | Omega-3 | |
|---|---|---|---|---|---|---|---|---|---|
| Factors | Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 | Factor 8 | Communality |
| C13:0 | 0.781 | −0.034 | −0.036 | 0.177 | −0.077 | −0.059 | −0.134 | −0.145 | 0.692 |
| C14:0iso | 0.877 | −0.172 | −0.051 | 0.096 | 0.063 | −0.080 | 0.147 | −0.060 | 0.847 |
| C15:0 | 0.849 | 0.030 | −0.122 | 0.021 | 0.017 | 0.181 | 0.000 | 0.033 | 0.772 |
| C16:0iso | 0.872 | −0.032 | −0.098 | −0.090 | −0.021 | 0.085 | 0.150 | −0.049 | 0.812 |
| C20:0 | 0.673 | −0.322 | 0.055 | −0.076 | −0.288 | 0.155 | 0.373 | −0.009 | 0.813 |
| C20:4 n6 | 0.795 | −0.201 | 0.078 | 0.155 | −0.237 | −0.066 | −0.183 | 0.241 | 0.854 |
| C24:0 | 0.686 | −0.073 | −0.146 | −0.235 | −0.066 | 0.145 | 0.355 | −0.008 | 0.734 |
| C22:5 n3 | 0.722 | −0.129 | −0.046 | −0.005 | 0.043 | −0.115 | −0.004 | 0.690 | 0.904 |
| C22:6 n3 | 0.714 | −0.277 | 0.040 | 0.016 | −0.044 | −0.227 | −0.219 | 0.669 | 0.825 |
| C8:0 | −0.088 | 0.754 | −0.142 | 0.239 | −0.015 | −0.350 | −0.164 | 0.052 | 0.806 |
| C10:0 | −0.132 | 0.942 | −0.164 | 0.040 | −0.130 | −0.063 | −0.111 | 0.013 | 0.966 |
| C12:0 | −0.172 | 0.916 | −0.132 | −0.029 | −0.140 | 0.176 | −0.089 | −0.001 | 0.945 |
| C14:0 | −0.182 | 0.686 | −0.067 | −0.326 | −0.211 | 0.426 | 0.047 | −0.037 | 0.845 |
| C18:1c9 | 0.302 | −0.603 | 0.173 | 0.214 | −0.092 | −0.006 | 0.056 | 0.295 | 0.629 |
| C18:1t6–8 | −0.218 | −0.201 | 0.814 | −0.115 | 0.321 | −0.247 | 0.012 | −0.105 | 0.938 |
| C18:1t9 | −0.282 | −0.168 | 0.771 | −0.109 | 0.408 | −0.190 | 0.087 | −0.044 | 0.927 |
| C18:1t10 | −0.097 | −0.112 | 0.809 | 0.045 | 0.141 | −0.103 | −0.178 | −0.085 | 0.748 |
| C18:1c12 | 0.285 | −0.151 | 0.684 | −0.273 | −0.145 | 0.065 | 0.006 | −0.006 | 0.672 |
| C18:1t15 | −0.093 | −0.081 | −0.663 | 0.397 | 0.129 | 0.041 | 0.271 | −0.163 | 0.731 |
| C10:1c9 | 0.012 | 0.466 | 0.167 | 0.638 | −0.035 | 0.045 | −0.235 | −0.110 | 0.723 |
| C14:1c9 | −0.145 | 0.085 | −0.245 | 0.637 | −0.048 | 0.468 | −0.182 | 0.130 | 0.765 |
| C18:1t11 | −0.325 | −0.082 | 0.240 | −0.109 | 0.822 | −0.211 | 0.053 | 0.025 | 0.906 |
| C18:2c9t11 | 0.074 | −0.153 | 0.168 | −0.029 | 0.910 | −0.071 | −0.217 | 0.122 | 0.953 |
| C16:0 | 0.028 | 0.162 | −0.068 | −0.335 | −0.369 | 0.680 | 0.100 | 0.076 | 0.633 |
| C16:1c9 | 0.120 | −0.024 | −0.191 | −0.032 | −0.082 | 0.660 | −0.086 | −0.127 | 0.518 |
| C18:0 | −0.119 | −0.316 | 0.032 | −0.084 | −0.074 | −0.204 | 0.689 | 0.034 | 0.517 |
| C4:0 | 0.468 | −0.178 | −0.107 | 0.227 | 0.054 | −0.314 | −0.663 | 0.115 | 0.597 |
| C6:0 | 0.158 | 0.180 | −0.034 | −0.216 | 0.018 | −0.585 | −0.622 | −0.052 | 0.629 |
| C20:5 n3 | 0.215 | −0.028 | −0.258 | 0.031 | 0.214 | −0.074 | 0.005 | 0.638 | 0.573 |
| C18:2n6 | 0.434 | 0.024 | 0.251 | 0.355 | −0.118 | 0.016 | 0.147 | 0.286 | 0.495 |
| C18:3n3 | 0.004 | −0.094 | 0.121 | −0.134 | 0.396 | −0.410 | 0.158 | 0.214 | 0.437 |
| C20:1c11 | −0.365 | 0.043 | 0.171 | 0.279 | 0.180 | 0.001 | 0.004 | −0.178 | 0.306 |
| C21:0 | 0.302 | −0.034 | −0.123 | −0.342 | 0.045 | 0.131 | 0.339 | −0.042 | 0.361 |
| Eigenvalue | 7.32 | 5.76 | 3.84 | 2.99 | 2.09 | 1.55 | 1.37 | 1.22 | |
| Variance explained | 22.18 | 17.45 | 11.62 | 9.07 | 6.34 | 4.69 | 4.15 | 3.70 | |
| Cumulative variance | 22.18 | 39.63 | 51.26 | 60.33 | 66.66 | 71.36 | 75.50 | 79.20 |
| Factors | Findings |
|---|---|
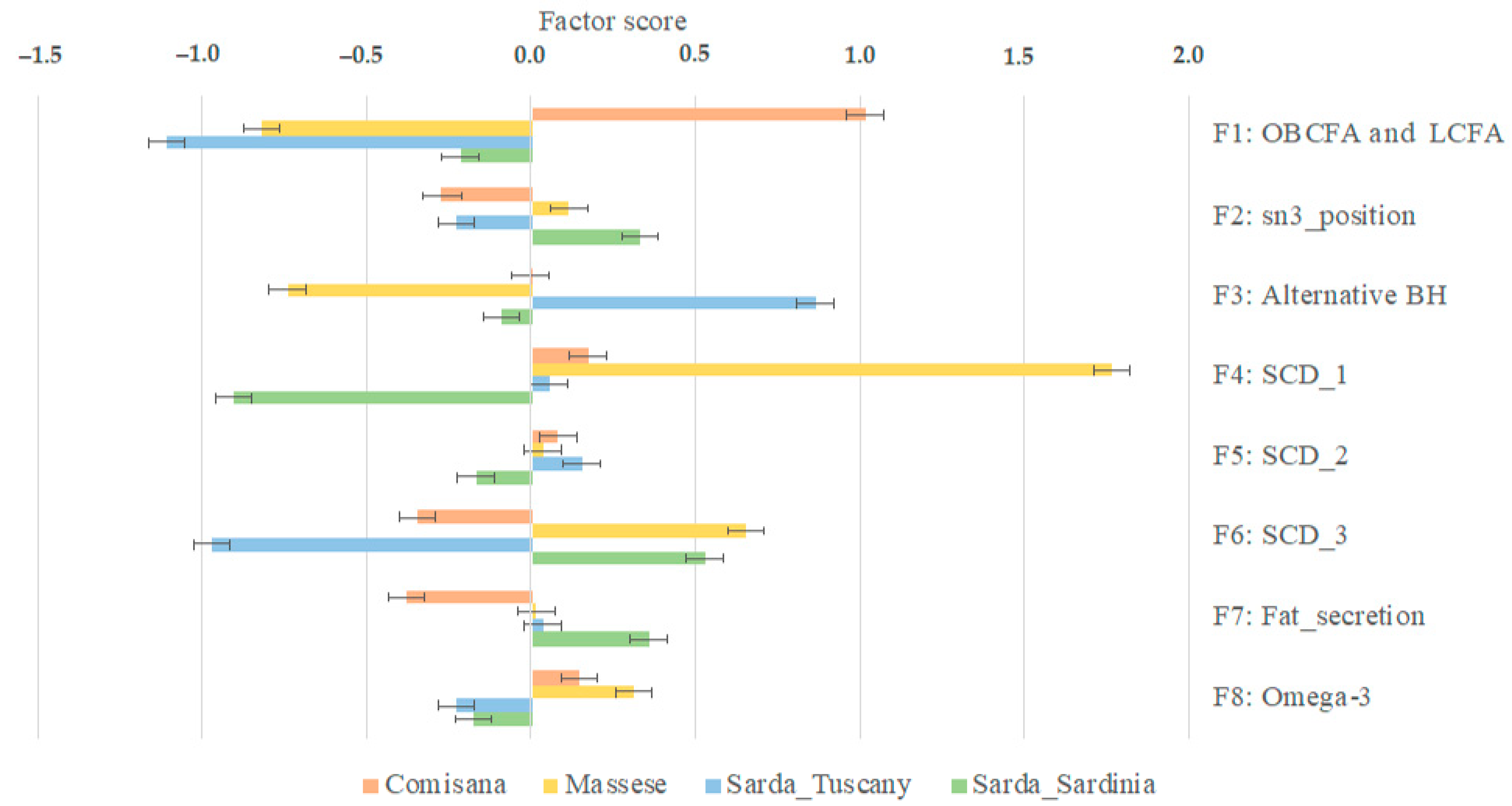
| Factor 1 = OBCFA and LCFA | Comisana was the only breed that showed positive scores |
| Factor 2 = sn3_position | Massese and Sarda_Sardinia showed positive scores, while Comisana and Sarda_Tuscany showed negative scores |
| Factor 3 = Alternative BH | Sarda_Tuscany was the only breed that showed positive scores |
| Factor 4 = SCD_1 | Sarda_Sardinia was the only breed that showed negative scores |
| Factor 5 = SCD_2 | Sarda_Sardinia was the only breed that showed negative scores |
| Factor 6 = SCD_3 | Massese and Sarda_Sardinia showed positive scores, while Comisana and Sarda_Tuscany showed negative scores |
| Factor 7 = Fat secretion | Comisana was the only breed that showed negative scores |
| Factor 8 = Omega-3 | Massese and Comisana showed positive scores, while Sarda_Sardinia and Sarda_Tuscany showed negative scores |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, G.; Palombo, V.; Serra, A.; Correddu, F.; D’Andrea, M.; Macciotta, N.P.P.; Mele, M. Study of the Fatty Acid Profile of Milk in Different Sheep Breeds: Evaluation by Multivariate Factorial Analysis. Animals 2022, 12, 722. https://doi.org/10.3390/ani12060722
Conte G, Palombo V, Serra A, Correddu F, D’Andrea M, Macciotta NPP, Mele M. Study of the Fatty Acid Profile of Milk in Different Sheep Breeds: Evaluation by Multivariate Factorial Analysis. Animals. 2022; 12(6):722. https://doi.org/10.3390/ani12060722
Chicago/Turabian StyleConte, Giuseppe, Valentino Palombo, Andrea Serra, Fabio Correddu, Mariasilvia D’Andrea, Nicolò Pietro Paolo Macciotta, and Marcello Mele. 2022. "Study of the Fatty Acid Profile of Milk in Different Sheep Breeds: Evaluation by Multivariate Factorial Analysis" Animals 12, no. 6: 722. https://doi.org/10.3390/ani12060722
APA StyleConte, G., Palombo, V., Serra, A., Correddu, F., D’Andrea, M., Macciotta, N. P. P., & Mele, M. (2022). Study of the Fatty Acid Profile of Milk in Different Sheep Breeds: Evaluation by Multivariate Factorial Analysis. Animals, 12(6), 722. https://doi.org/10.3390/ani12060722








