The Mechanisms of Fur Development and Color Formation in American Mink Revealed Using Comparative Transcriptomics
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. RNA Quantification and Qualification
2.3. Library Preparation for Transcriptome Sequencing
2.4. Clustering and Sequencing
2.5. Quality Control
2.6. Read Mapping to the Reference Genome
2.7. Novel Transcripts Prediction
2.8. Quantification of Gene Expression Level
2.9. Differential Expression Analysis
2.10. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
2.11. Verification of Differentially Expressed Genes by qPCR
3. Results
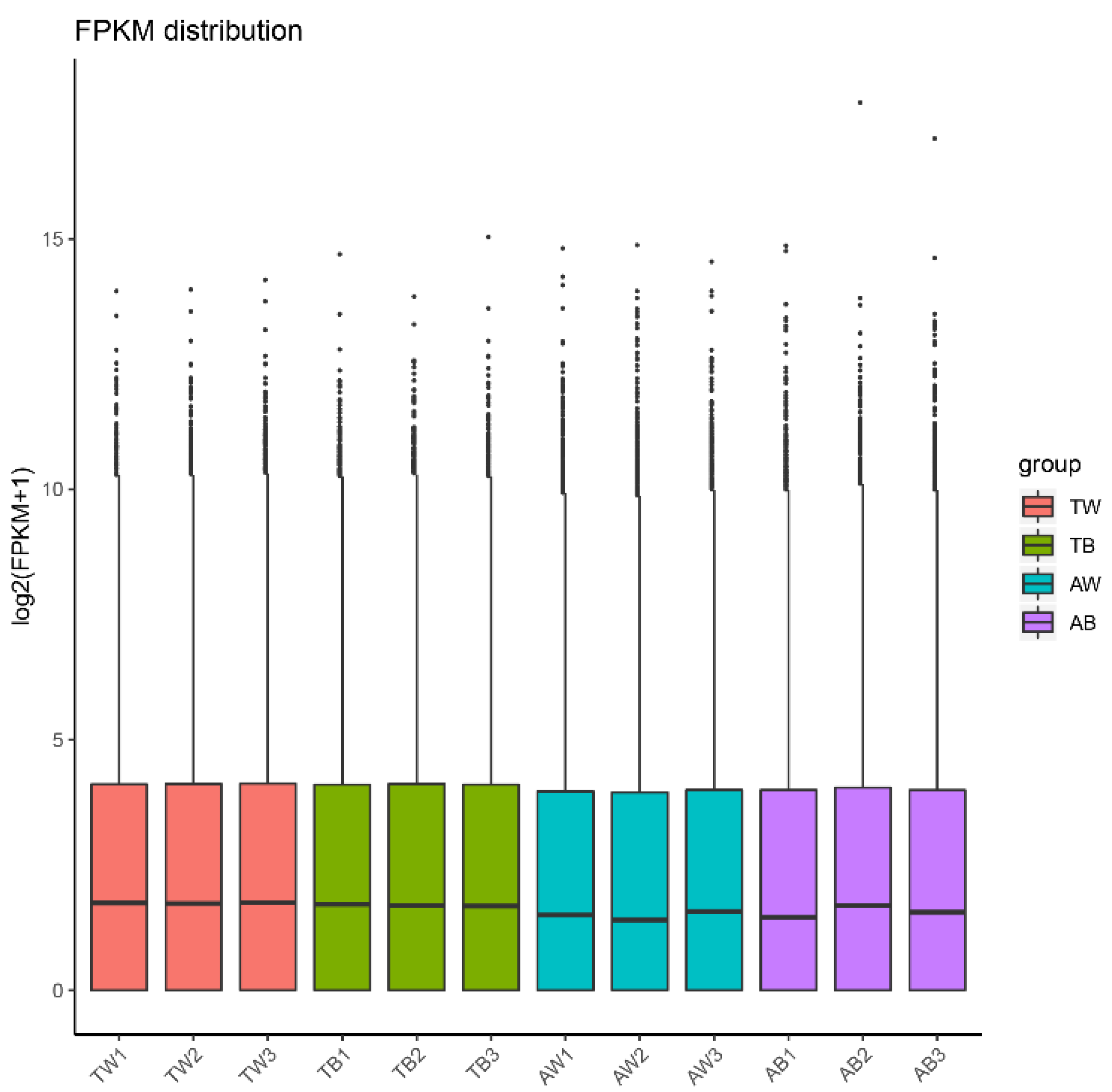
3.1. Data Quality Control and Novel Gene Prediction
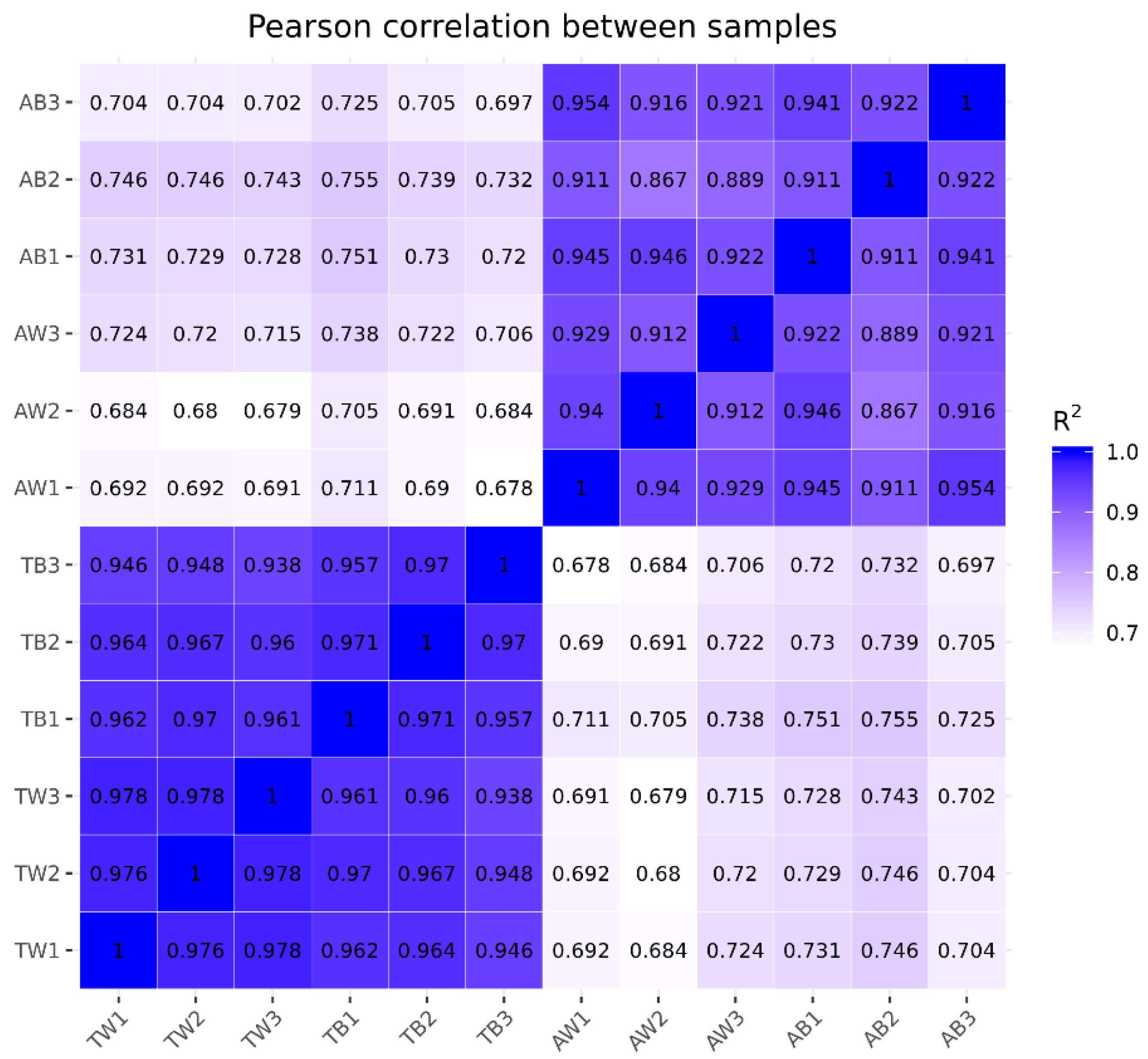
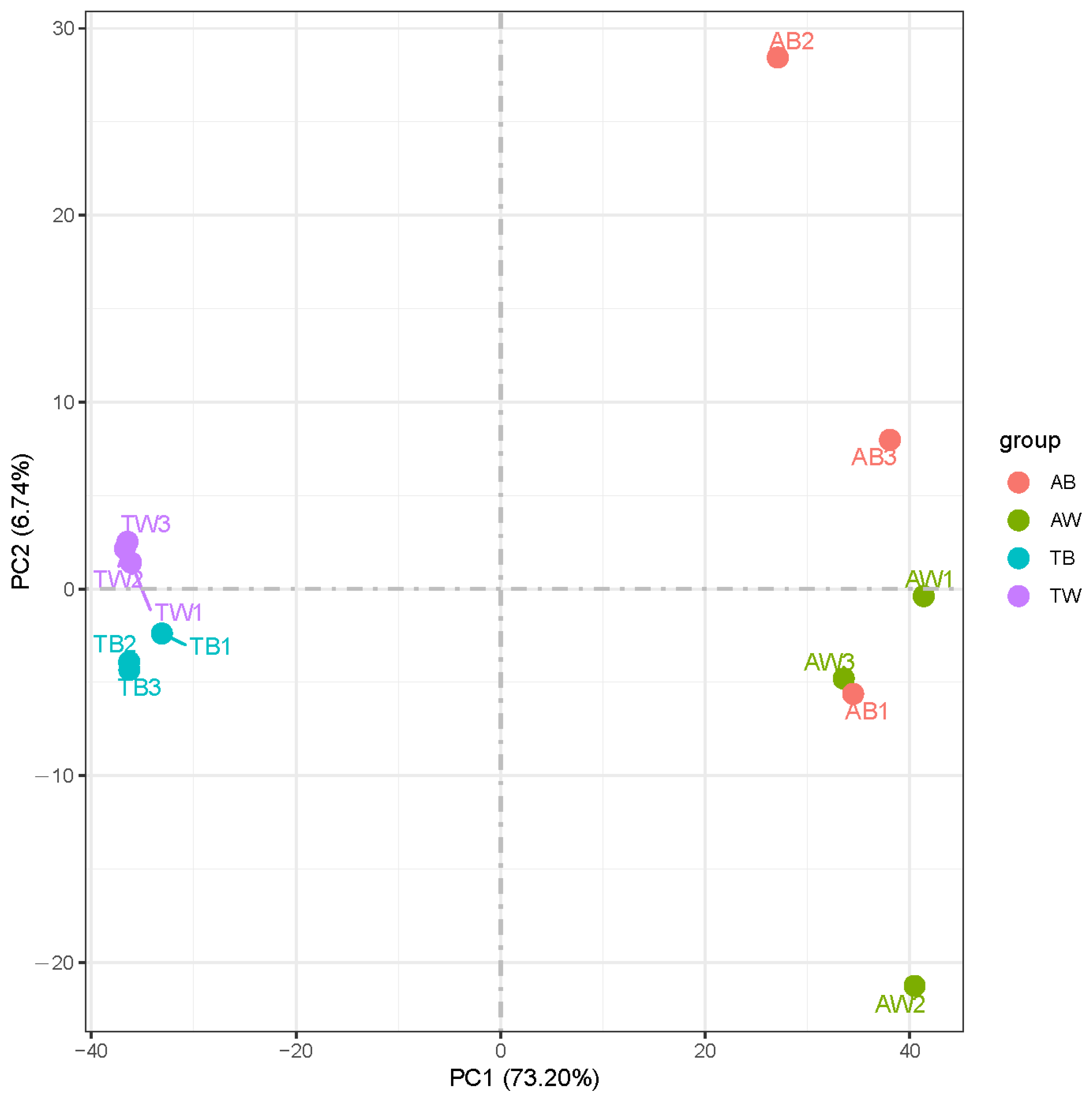
3.2. Quantitative Analysis of Gene Expression
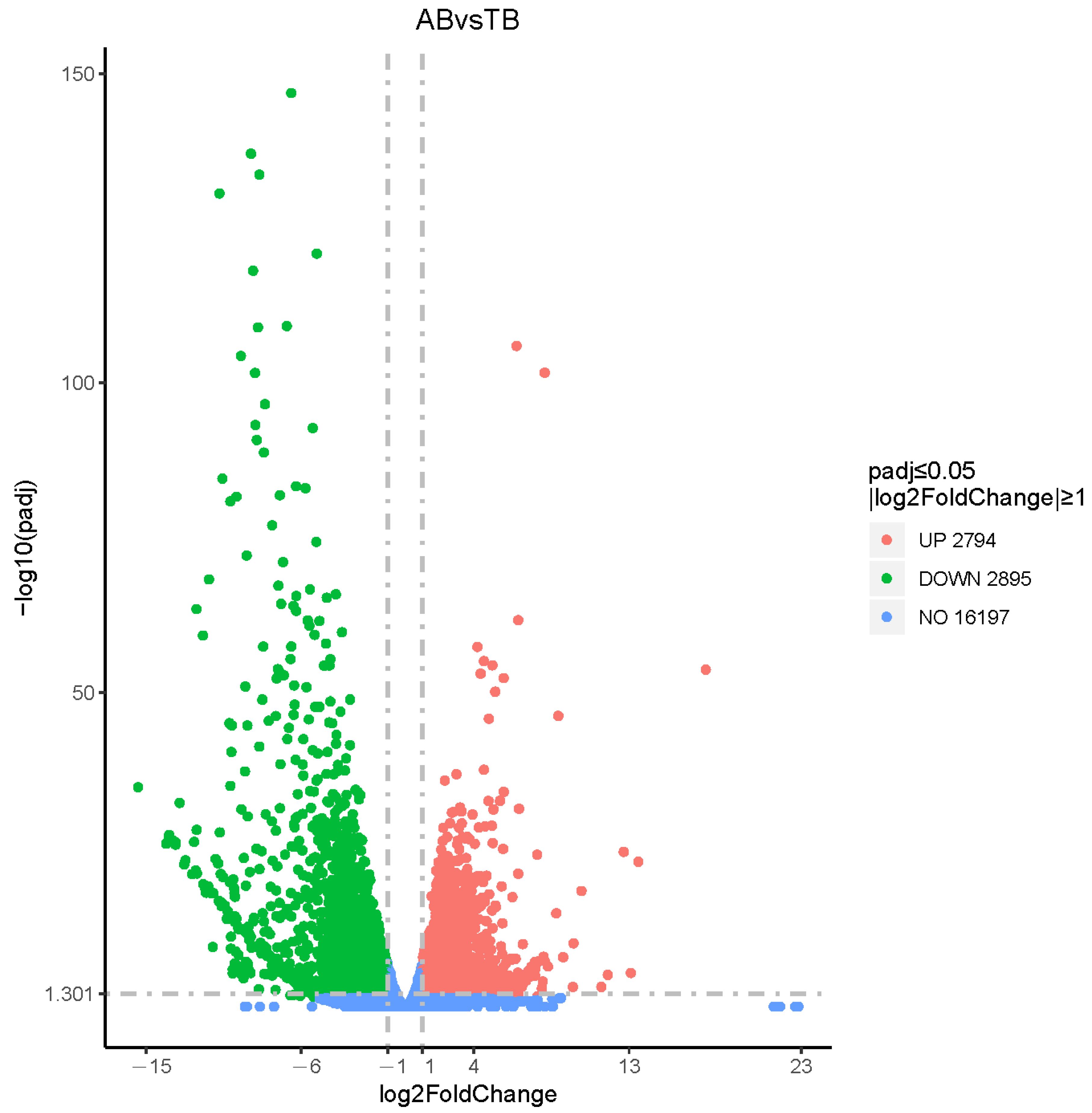
3.3. Differentially Expressed Genes (DEGs) in American Mink Skin
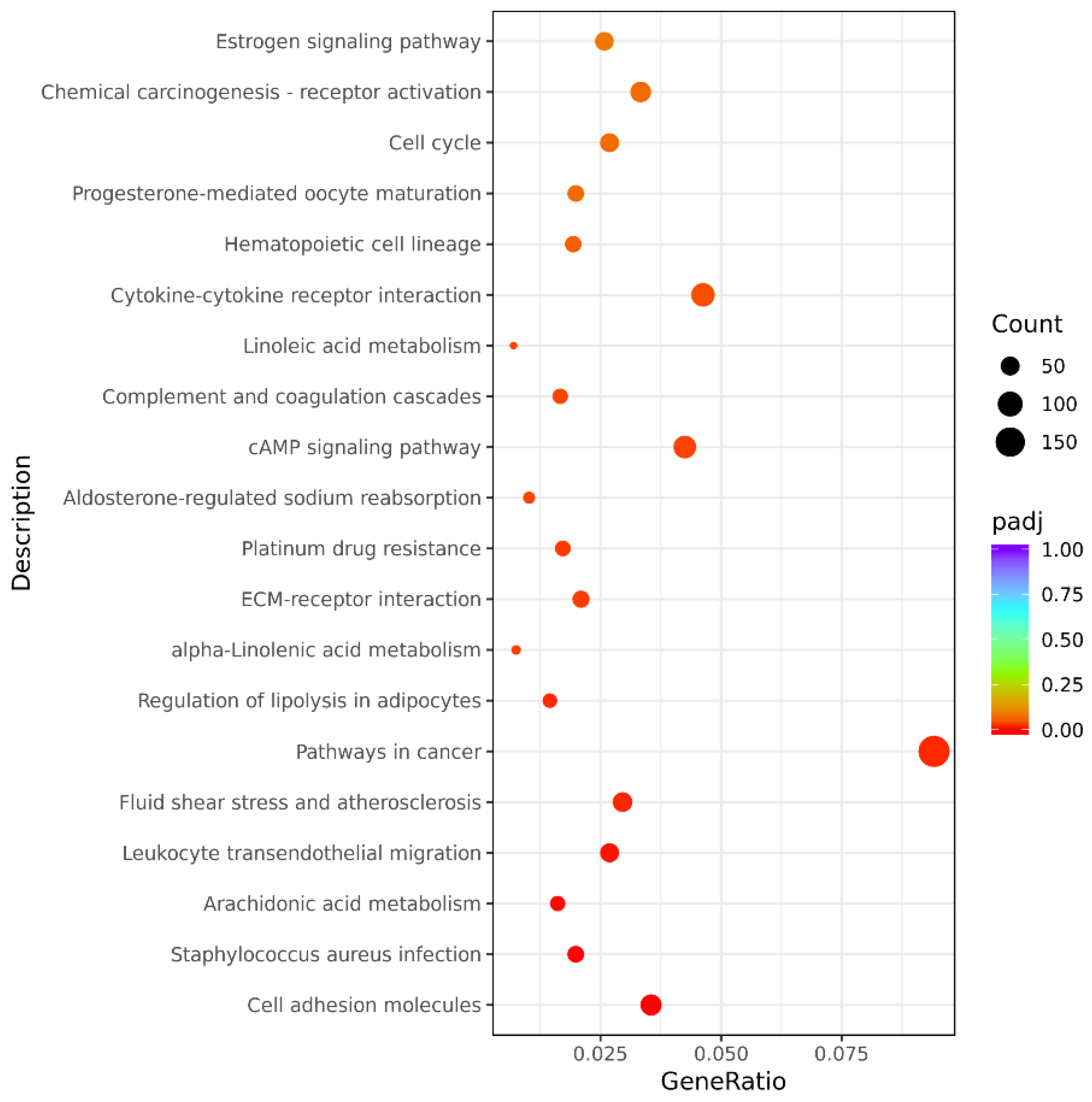
3.4. Analysis of GO and KEGG Pathways
3.5. qPCR Validation of Differential Gene Expression among Groups of American Mink
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manakhov, A.D.; Andreeva, T.V.; Trapezov, O.V.; Kolchanov, N.A.; Rogaev, E.I. Genome analysis identifies the mutant genes for common industrial Silverblue and Hedlund white coat colours in American mink. Sci. Rep. 2019, 9, 4581. [Google Scholar] [CrossRef] [PubMed]
- Winfield, I.J. Vertebrates: Fish, Amphibians, Reptiles, Birds, Mammals; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Joergensen, G. Mink Production; Scientifur: Hilleroed, Denmark, 1985. [Google Scholar]
- Falconer, D.S.; Mackay, T.F. Introduction to Quantitative Genetics; Benjamin-Cummings: San Francisco, CA, USA, 1960. [Google Scholar]
- Anistoroaei, R.; Krogh, A.K.; Christensen, K. A frameshift mutation in the LYST gene is responsible for the Aleutian color and the associated Chédiak-Higashi syndrome in American mink. Anim. Genet. 2013, 44, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Anistoroaei, R.; Fredholm, M.; Christensen, K.; Leeb, T. Albinism in the American mink (Neovison vison) is associated with a tyrosinase nonsense mutation. Anim. Genet. 2008, 39, 645–648. [Google Scholar] [CrossRef]
- Cirera, S.; Markakis, M.N.; Kristiansen, T.; Vissenberg, K.; Fredholm, M.; Christensen, K.; Anistoroaei, R. A large insertion in intron 2 of the TYRP1 gene associated with American Palomino phenotype in American mink. Mamm. Genome 2016, 27, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Manakhov, A.D.; Mintseva, M.Y.; Andreev, I.A.; Uralsky, L.I.; Andreeva, T.V.; Trapezov, O.V.; Rogaev, E.I. Genome analysis of American minks reveals link of mutations in Ras-related protein-38 gene to Moyle brown coat phenotype. Sci. Rep. 2020, 10, 15876. [Google Scholar] [CrossRef]
- Benkel, B.F.; Rouvinen-Watt, K.; Farid, H.; Anistoroaei, R. Molecular characterization of the Himalayan mink. Mamm. Genome 2009, 20, 256–259. [Google Scholar] [CrossRef]
- Li, Y.C.; He, D.Q.; Ma, Y.H.; Ma, Q.; Ding, W.; Chen, Y.H.; Zhang, M.; Luo, F.; Chen, L.Y.; Wang, J.K.; et al. Skin transcriptome analysis identifies the key genes underlying fur development in Chinese Tan sheep in the birth and Er-mao periods. Gene 2022, 820, 146257. [Google Scholar] [CrossRef]
- Polakis, P. Wnt signaling and cancer. Genes Dev. 2000, 14, 1837–1851. [Google Scholar] [CrossRef]
- Mikkola, M.L. Genetic basis of skin appendage development. Semin. Cell Dev. Biol. 2007, 18, 225–236. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Smyth, G.K.; Wei, S. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal RNA-Seq quantification. Comput. Sci. 2016, 34, 525–527. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Lamoreux, M.L.; Wakamatsu, K.; Ito, S. Interaction of Major Coat Color Gene Functions in Mice as Studied by Chemical Analysis of Eumelanin and Pheomelanin. Pigment. Cell Res. 2001, 14, 23–31. [Google Scholar] [CrossRef]
- Baxter, L.L.; Watkins-Chow, D.E.; Pavan, W.J.; Loftus, S.K. A curated gene list for expanding the horizons of pigmentation biology. Pigment. Cell Melanoma Res. 2019, 32, 348–358. [Google Scholar] [CrossRef]
- Gu, L.-H.; Coulombe, P.A. Keratin expression provides novel insight into the morphogenesis and function of the companion layer in hair follicles. J. Investig. Dermatol. 2007, 127, 1061–1073. [Google Scholar] [CrossRef]
- Parry, A.L.; Nixon, A.J.; Craven, A.J.; Pearson, A.J. The Microanatomy, Cell Replication, and Keratin Gene Expression of Hair Follicles during a Photoperiod-lnduced Growth Cycle in Sheep. Acta Anat. 1995, 154, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Gordon, S.W.; Nixon, A.J.; Bawden, C.S.; Rogers, M.A.; Wildermoth, J.E.; Maqbool, N.J.; Pearson, A.J. Expression patterns of keratin intermediate filament and keratin associated protein genes in wool follicles. Differentiation 2009, 77, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Millar, S.E.; Willert, K.; Salinas, P.C.; Roelink, H.; Nusse, R.; Sussman, D.J.; Barsh, G.S. WNT signaling in the control of hair growth and structure. Dev. Biol. 1999, 207, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-S.; Francis-West, P.H.; Widelitz, R.B.; Jiang, T.-X.; Ting-Berreth, S.; Tickle, C.; Wolpert, L.; Chuong, C.-M. Local inhibitory action of BMPs and their relationships with activators in feather formation: Implications for periodic patterning. Dev. Biol. 1998, 196, 11–23. [Google Scholar] [CrossRef]
- Rao, R.K.; Philipps, A.F.; Williams, C.S.; McCracken, D.M.; Koldovsky, O. Luminal stability of insulin-like growth factors I and II in developing rat gastrointestinal tract. J. Pediatr. Gastroenterol. Nutr. 1998, 26, 179–185. [Google Scholar] [CrossRef]
- Weger, N.; Schlake, T. Igf-I signalling controls the hair growth cycle and the differentiation of hair shafts. J. Investig. Dermatol. 2005, 125, 873–882. [Google Scholar] [CrossRef]
- Coulombe, P.A.; Omary, M.B. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002, 14, 110–122. [Google Scholar] [CrossRef]
- Steinert, P.M.; Torchia, D.R.; Mack, J.W. Structural Features of Keratin Intermediate Filaments. In The Biology of Wool and Hair; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar]
- Moll, R.; Divo, M.; Langbein, L. The human keratins: Biology and pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef]
- Cadieu, E.; Neff, M.W.; Quignon, P.; Walsh, K.; Chase, K.; Parker, H.G.; Vonholdt, B.M.; Rhue, A.; Boyko, A.; Byers, A.; et al. Coat variation in the domestic dog is governed by variants in three genes. Science 2009, 326, 150–153. [Google Scholar] [CrossRef]
- Gandolfi, B.; Outerbridge, C.A.; Beresford, L.G.; Myers, J.A.; Pimentel, M.; Alhaddad, H.; Grahn, J.C.; Grahn, R.A.; Lyons, L.A. The naked truth: Sphynx and Devon Rex cat breed mutations in KRT71. Mamm. Genome 2010, 21, 509–515. [Google Scholar] [CrossRef]
- Runkel, F.; Klaften, M.; Koch, K.; Bhnert, V.; Büssow, H.; Fuchs, H.; Franz, T.; de Angelis, M.H. Morphologic and molecular characterization of two novel Krt71 (Krt2-6g) mutations: Krt71rco12 and Krt71rco13. Mamm. Genome 2006, 17, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Lemke, J.R.; Kernland-Lang, K.; Hrtnagel, K.; Itin, P. Monogenic Human Skin Disorders. Dermatology 2014, 229, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, J.; Langbein, L.; Rogers, M.A.; Winter, H. Hair follicle-specific keratins and their diseases. Exp. Cell Res. 2007, 313, 2010–2020. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-D.; Irwin, D.M.; Zhang, Y.-P. Molecular evolution of the keratin associated protein gene family in mammals, role in the evolution of mammalian hair. BMC Evol. Biol. 2009, 9, 213. [Google Scholar] [CrossRef]
- Khan, I.; Maldonado, E.; Vasconcelos, V.; O’Brien, S.J.; Johnson, W.E.; Antunes, A. Mammalian keratin associated proteins (KRTAPs) subgenomes: Disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genom. 2014, 15, 779. [Google Scholar] [CrossRef]
- Zhou, H.; Gong, H.; Li, S.; Luo, Y.; Hickford, J. A 57-bp deletion in the ovine KAP6-1 gene affects wool fibre diameter. J. Anim. Breed. Genet. 2015, 132, 301–307. [Google Scholar] [CrossRef]
- Li, S.; Zhou, H.; Gong, H.; Zhao, F.; Wang, J.; Liu, X.; Luo, Y.; Hickford, J.G.H. Identification of the Ovine Keratin-Associated Protein 22-1 (KAP22-1) Gene and Its Effect on Wool Traits. Genes 2017, 8, 27. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Dyer, J.M.; Hickford, J.G.H. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011, 38, 5429–5433. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Hickford, J.G.H. Polymorphism of the ovine keratin-associated protein 1-4 gene (KRTAP1-4). Mol. Biol. Rep. 2010, 37, 3377–3380. [Google Scholar] [CrossRef]
- Rogers, M.A.; Langbein, L.; Praetzel-Wunder, S.; Winter, H.; Schweizer, J. Human hair keratin-associated proteins (KAPs). Int. Rev. Cytol. 2006, 251, 209–263. [Google Scholar]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Itoh, F.; Watabe, T.; Miyazono, K. Roles of TGF-β family signals in the fate determination of pluripotent stem cells. Semin. Cell Dev. Biol. 2014, 32, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Dimitriou, R.; Giannoudis, P.V. Discovery and development of BMPs. Injury 2005, 36, S28–S33. [Google Scholar] [CrossRef] [PubMed]
- Rosso, F.; Giordano, A.; Barbarisi, M.; Barbarisi, A. From cell-ECM interactions to tissue engineering. J. Cell. Physiol. 2004, 199, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Comoglio, P.M.; Boccaccio, C.; Trusolino, L. Interactions between growth factor receptors and adhesion molecules: Breaking the rules. Curr. Opin. Cell Biol. 2003, 15, 565–571. [Google Scholar] [CrossRef]
- Parmeggiani, A. GTPases—Molecular switches of cellular signaling pathways. Trends Biochem. Sci. 2001, 26, 76. [Google Scholar] [CrossRef]
- Jimbow, K.; Park, J.S.; Kato, F.; Hirosaki, K.; Toyofuku, K.; Hua, C.; Yamashita, T. Assembly, Target-Signaling and Intracellular Transport of Tyrosinase Gene Family Proteins in the Initial Stage of Melanosome Biogenesis. Pigment. Cell Res. 2000, 13, 222–229. [Google Scholar] [CrossRef]
- Hirobe, T.; Wakamatsu, K.; Ito, S. Excess Tyrosine Stimulates Eumelanin and Pheomelanin Synthesis in Cultured Slaty Melanocytes from Neonatal Mouse Epidermis. Zool. Sci. 2007, 24, 209–217. [Google Scholar] [CrossRef]
- Makpol, S.; Jam, F.A.; Rahim, N.A.; Khor, S.C.; Ismail, Z.; Yusof, Y.A.M.; Ngah, W.Z.W. Comparable down-regulation of TYR, TYRP1 and TYRP2 genes and inhibition of melanogenesis by tyrostat, tocotrienol-rich fraction and tocopherol in human skin melanocytes improves skin pigmentation. Clin. Ter. 2014, 165, e39–e45. [Google Scholar]
- Marçon, C.R.; Maia, M. Albinism: Epidemiology, genetics, cutaneous characterization, psychosocial factors. An. Bras. Dermatol. 2019, 94, 503–520. [Google Scholar] [CrossRef]
- Shakil, M.; Harlalka, G.V.; Ali, S.; Lin, S.; D’Atri, I.; Hussain, S.; Nasir, A.; Shahzad, M.A.; Ullah, M.I.; Self, J.E.; et al. Tyrosinase (TYR) gene sequencing and literature review reveals recurrent mutations and multiple population founder gene mutations as causative of oculocutaneous albinism (OCA) in Pakistani families. Eye 2019, 33, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Aigner, B.; Besenfelder, U.; Müller, M.; Brem, G. Tyrosinase gene variants in different rabbit strains. Mamm. Genome 2000, 11, 700–702. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, S.M.; Berryere, T.G.; Ciobanu, D.C.; Mileham, A.J.; Schmidtz, B.H.; Fredholm, M. A form of albinism in cattle is caused by a tyrosinase frameshift mutation. Mamm. Genome 2004, 15, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Blaszczyk, W.M.; Arning, L.; Hoffmann, K.-P.; Epplen, J.T. A Tyrosinase missense mutation causes albinism in the Wistar rat. Pigment. Cell Res. 2005, 18, 144–145. [Google Scholar] [CrossRef]
- Imes, D.L.; Geary, L.A.; Grahn, R.A.; Lyons, L.A. Albinism in the domestic cat (Felis catus) is associated with a tyrosinase (TYR) mutation. Anim. Genet. 2006, 37, 175–178. [Google Scholar] [CrossRef]
- Blaszczyk, W.M.; Distler, C.; Dekomien, G.; Arning, L.; Hoffmann, K.-P.; Epplen, J.T. Identification of a tyrosinase (TYR) exon 4 deletion in albino ferrets (Mustela putorius furo). Anim. Genet. 2007, 38, 421–423. [Google Scholar] [CrossRef]
- King, R.A.; Pietsch, J.; Fryer, J.P.; Savage, S.; Brott, M.J.; Russell-Eggitt, I.; Summers, C.G.; Oetting, W.S. Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): Definition of the phenotype. Hum. Genet. 2003, 113, 502–513. [Google Scholar] [CrossRef]
- Wan, P.; Hu, Y.; He, L. Regulation of melanocyte pivotal transcription factor MITF by some other transcription factors. Mol. Cell. Biochem. 2011, 354, 241–246. [Google Scholar] [CrossRef]
- Hida, T.; Wakamatsu, K.; Sviderskaya, E.V.; Donkin, A.J.; Montoliu, L.; Lynn Lamoreux, M.; Yu, B.; Millhauser, G.L.; Ito, S.; Barsh, G.S.; et al. Agouti protein, mahogany, and attractin in pheomelanogenesis and melanoblast-like alteration of melanocytes: A cAMP-independent pathway. Pigment. Cell Melanoma Res. 2009, 22, 623–634. [Google Scholar] [CrossRef]
- Nasti, T.H.; Timares, L. MC1R, Eumelanin and Pheomelanin: Their role in determining the susceptibility to skin cancer. Photochem. Photobiol. 2015, 91, 188–200. [Google Scholar] [CrossRef]
- Suzuki, I.; Tada, A.; Ollmann, M.M.; Barsh, G.S.; Im, S.; Lamoreux, M.L.; Hearing, V.J.; Nordlund, J.J.; Abdel-Malek, Z.A. Agouti signaling protein inhibits melanogenesis and the response of human melanocytes to α-melanotropin. J. Investig. Dermatol. 1997, 108, 838–842. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, X.; Zhou, R.; Li, L.; Yang, Q. Bioinformatics analysis of complete coding regions of TYRP2 gene among 21 species. J. Henan Agric. Sci. 2011, 40, 144–148. [Google Scholar]


| Sample | Raw-Reads | Clean-Reads | Clean-Bases | Q20 | Q30 | GC-pct |
|---|---|---|---|---|---|---|
| AB1 | 45,947,094 | 45,190,978 | 6.78 G | 97.7 | 93.57 | 52.55 |
| AB2 | 43,685,582 | 42,895,824 | 6.43 G | 97.83 | 93.81 | 52.43 |
| AB3 | 46,277,030 | 45,357,284 | 6.8 G | 97.82 | 93.83 | 52.28 |
| AW1 | 44,938,430 | 44,155,634 | 6.62 G | 97.73 | 93.69 | 52.94 |
| AW2 | 48,298,078 | 47,408,254 | 7.11 G | 97.88 | 93.94 | 51.17 |
| AW3 | 46,758,560 | 46,088,186 | 6.91 G | 97.84 | 93.87 | 51.52 |
| TB1 | 43,634,474 | 42,882,980 | 6.43 G | 97.76 | 93.71 | 52.22 |
| TB2 | 47,393,846 | 46,084,054 | 6.91 G | 98.24 | 94.89 | 51.14 |
| TB3 | 46,278,052 | 44,685,668 | 6.7 G | 98.18 | 94.64 | 49.18 |
| TW1 | 44,342,806 | 43,591,538 | 6.54 G | 97.68 | 93.56 | 52.31 |
| TW2 | 44,828,808 | 43,975,908 | 6.6 G | 97.71 | 93.62 | 52.77 |
| TW3 | 46,740,826 | 45,858,686 | 6.88 G | 97.75 | 93.72 | 52.91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhou, S.; Liu, G.; Lyu, T.; Shi, L.; Dong, Y.; He, S.; Zhang, H. The Mechanisms of Fur Development and Color Formation in American Mink Revealed Using Comparative Transcriptomics. Animals 2022, 12, 3088. https://doi.org/10.3390/ani12223088
Wang L, Zhou S, Liu G, Lyu T, Shi L, Dong Y, He S, Zhang H. The Mechanisms of Fur Development and Color Formation in American Mink Revealed Using Comparative Transcriptomics. Animals. 2022; 12(22):3088. https://doi.org/10.3390/ani12223088
Chicago/Turabian StyleWang, Lidong, Shengyang Zhou, Guangshuai Liu, Tianshu Lyu, Lupeng Shi, Yuehuan Dong, Shangbin He, and Honghai Zhang. 2022. "The Mechanisms of Fur Development and Color Formation in American Mink Revealed Using Comparative Transcriptomics" Animals 12, no. 22: 3088. https://doi.org/10.3390/ani12223088
APA StyleWang, L., Zhou, S., Liu, G., Lyu, T., Shi, L., Dong, Y., He, S., & Zhang, H. (2022). The Mechanisms of Fur Development and Color Formation in American Mink Revealed Using Comparative Transcriptomics. Animals, 12(22), 3088. https://doi.org/10.3390/ani12223088





