Oral Administration of a Phage Cocktail to Reduce Salmonella Colonization in Broiler Gastrointestinal Tract—A Pilot Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains Used and Culture Conditions
2.2. Phage Lysate Preparation, Host Range Determination, and Efficiency of Plating
2.3. Phage Cocktail Development and Phage Efficacy Test In Vitro
2.4. Study of Phage-Resistance Development in Salmonella upon Phage Cocktail Treatment
2.5. Phage Survivability in Simulated Chicken Gastrointestinal Conditions
2.6. Evaluation of Phage Cytotoxicity in Cell Lines
2.7. Efficacy of a Phage Cocktail in Reducing Salmonella Populations in a Broiler Gastrointestinal Tract
2.8. Sampling Method and Sample Examination
2.9. Statistical Analysis
3. Results
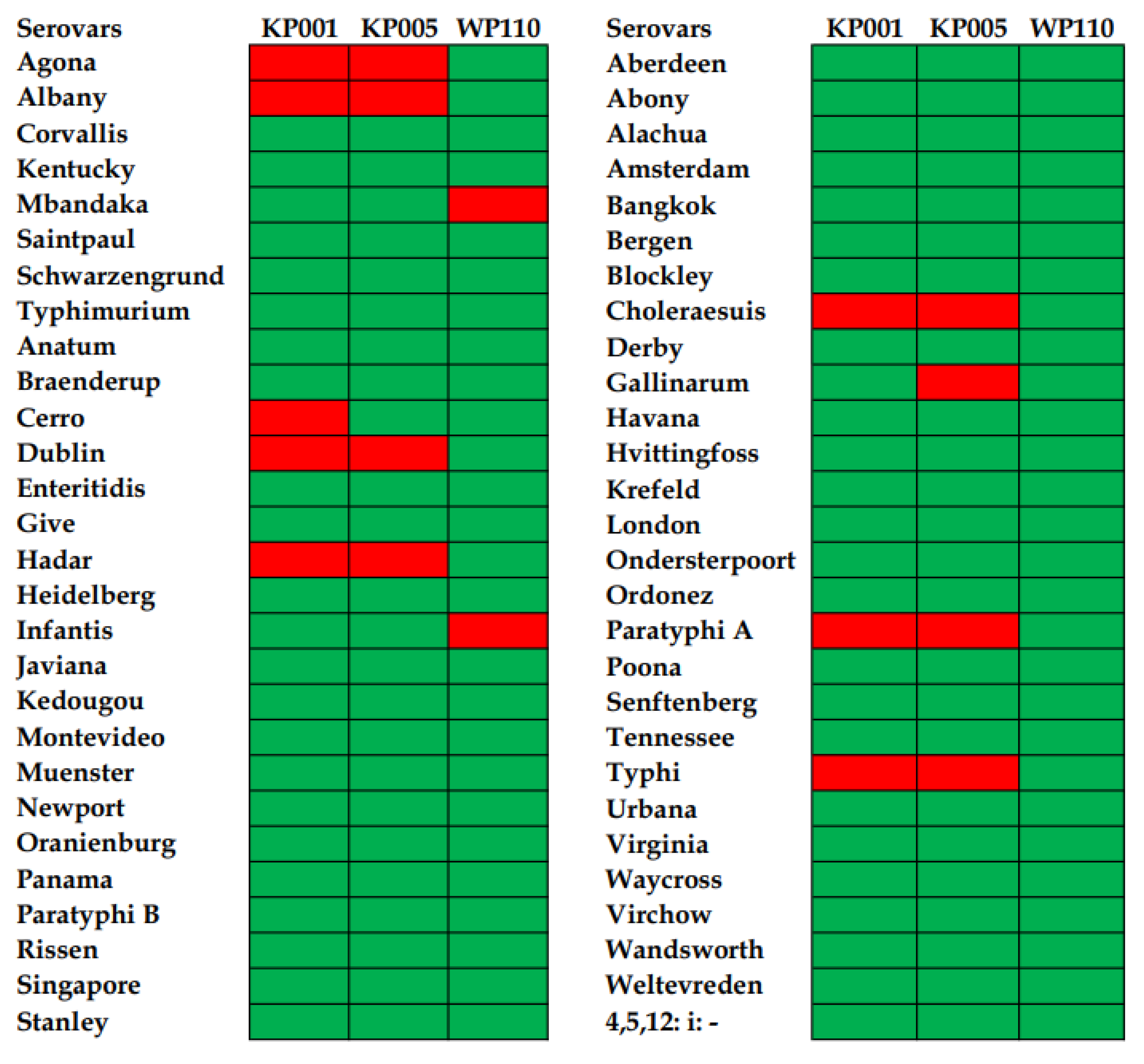
3.1. Phage Host Range against Diverse Salmonella Serovars and Efficiency of Plating
3.2. Evaluation of a Phage Cocktail Targeting Salmonella In Vitro
3.3. Study of Phage-Resistance Development in Salmonella after Treatment with Phage Cocktail
3.4. Phage Survivability in Simulated Chicken Gastrointestinal Conditions
3.5. Evaluation of Phage Cytotoxicity in Cell Lines
3.6. Reduction of Salmonella in the Broiler Gastrointestinal Tract after Phage Cocktail Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Popa, G.L.; Papa, M.I. Salmonella spp. infection-a continuous threat worldwide. Germs 2021, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). European Centre for Disease Prevention and Control (ECDC). The European Union one health 2019 zoonoses report. EFSA J. 2021, 19, e6406. [Google Scholar]
- Fagbamila, I.O.; Barco, L.; Mancin, M.; Kwaga, J.; Ngulukun, S.S.; Zavagnin, P.; Lettini, A.A.; Lorenzetto, M.; Abdu, P.A.; Kabir, J.; et al. Salmonella serovars and their distribution in Nigerian commercial chicken layer farms. PLoS ONE 2017, 12, e0173097. [Google Scholar] [CrossRef] [PubMed]
- Djeffal, S.; Mamache, B.; Elgroud, R.; Hireche, S.; Bouaziz, O. Prevalence and risk factors for Salmonella spp. contamination in broiler chicken farms and slaughterhouses in the northeast of Algeria. Vet. World 2018, 11, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Eguale, T. Non-typhoidal Salmonella serovars in poultry farms in central Ethiopia: Prevalence and antimicrobial resistance. BMC Vet. Res. 2018, 14, 217. [Google Scholar] [CrossRef] [PubMed]
- Witkowska, D.; Kuncewicz, M.; Źebrowska, J.P.; Sobczak, J.; Sowińska, J. Prevalence of Salmonella spp. in broiler chicken flocks in northern Poland in 2014–2016. Ann. Agric. Environ. Med. 2018, 25, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Pelyuntha, W.; Ngasaman, R.; Yingkajorn, M.; Chukiatsiri, K.; Benjakul, S.; Vongkamjan, K. Isolation and characterization of potential Salmonella phages targeting multidrug-resistant and major serovars of Salmonella derived from broiler production chain in Thailand. Front. Microbiol. 2021, 12, 662461. [Google Scholar] [CrossRef] [PubMed]
- Zha, L.; Garrett, S.; Sun, J. Salmonella infection in chronic inflammation and gastrointestinal cancer. Diseases 2019, 7, 28. [Google Scholar] [CrossRef]
- Clavijo, V.; Baquero, D.; Hernandez, S.; Farfan, J.C.; Arias, J.; Arévalo, A.; Donado-Godoy, P.; Vives-Flores, M. Phage cocktail SalmoFREE® reduces Salmonella on a commercial broiler farm. Poult. Sci. 2019, 98, 5054–5063. [Google Scholar] [CrossRef]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, food safety and food handling practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef]
- Van Immerseel, F.; De Zutter, L.; Houf, K.; Pasmans, F.; Haesebrouck, F.; Ducatelle, R. Strategies to control Salmonella in the broiler production chain. Worlds Poult. Sci. J. 2019, 65, 367–392. [Google Scholar] [CrossRef]
- Gu, G.; Strawn, L.K.; Oryang, D.O.; Zheng, J.; Reed, E.A.; Ottesen, A.R.; Bell, R.L.; Chen, Y.; Duret, S.; Ingram, D.T.; et al. Agricultural practices influence Salmonella contamination and survival in pre-harvest tomato production. Front. Microbiol. 2018, 9, 2451. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial resistance: Its surveillance, impact, and alternative management strategies in dairy animals. Front. Vet. Sci. 2018, 4, 237. [Google Scholar] [CrossRef] [PubMed]
- Agyare, C.; Boamah, V.E.; Zumbi, C.N.; Osei, F.B. Antibiotic use in poultry production and its effects on bacterial resistance. In Antimicrobial Resistance—A Global Threat; Kumar, Y., Ed.; IntechOpen Limited: London, UK, 2018; pp. 33–51. [Google Scholar]
- Andrew Selaledi, L.; Mohammed Hassan, Z.; Manyelo, T.G.; Mabelebele, M. The current status of the alternative use to antibiotics in poultry production: An African perspective. Antibiotics 2020, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial resistance in bacterial poultry pathogens: A review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef]
- Mehdi, Y.; Létourneau-Montminy, M.P.; Gaucher, M.L.; Chorfi, Y.; Suresh, G.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of antibiotics in broiler production: Global impacts and alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef]
- Diarra, M.S.; Silversides, F.G.; Diarrassouba, F.; Pritchard, J.; Masson, L.; Brousseau, R.; Bonnet, C.; Delaquis, P.; Bach, S.; Skura, B.J.; et al. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and Enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolates. Appl. Environ. Microbiol. 2007, 73, 6566–6576. [Google Scholar]
- Cuong, N.V.; Kiet, B.T.; Hien, V.B.; Truong, B.D.; Phu, D.H.; Thwaites, G.; Choisy, M.; Carrique-Mas, J. Antimicrobial use through consumption of medicated feeds in chicken flocks in the Mekong Delta of Vietnam: A three-year study before a ban on antimicrobial growth promoters. PLoS ONE 2021, 16, e0250082. [Google Scholar] [CrossRef]
- Pelyuntha, W.; Vongkamjan, K. Combined effects of Salmonella phage cocktail and organic acid for controlling Salmonella Enteritidis in chicken meat. Food Control. 2022, 133, 108653. [Google Scholar] [CrossRef]
- Żbikowska, K.; Michalczuk, M.; Dolka, B. The use of bacteriophages in the poultry industry. Animals 2020, 10, 872. [Google Scholar] [CrossRef] [PubMed]
- Ferriol-González, C.; Domingo-Calap, P. Phage therapy in livestock and companion animals. Antibiotics 2021, 10, 559. [Google Scholar] [CrossRef]
- Shan, J.; Ramachandran, A.; Thanki, A.M.; Vukusic, F.B.; Barylski, J.; Clokie, M.R. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 2018, 8, 5091. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Gonçalves, G.A.M.; Donato, T.C.; Baptista, A.A.S.; de Oliveira Corrêa, I.M.; Garcia, K.C.O.D.; Andreatti Filho, R.L. Bacteriophage-induced reduction in Salmonella Enteritidis counts in the crop of broiler chickens undergoing preslaughter feed withdrawal. Poult. Sci. 2014, 93, 216–220. [Google Scholar] [CrossRef]
- Fiorentin, L.; Vieira, N.D.; Barioni Jr, W. Oral treatment with bacteriophages reduces the concentration of Salmonella Enteritidis PT4 in caecal contents of broilers. Avian Pathol. 2005, 34, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Petsong, K.; Benjakul, S.; Chaturongakul, S.; Switt, A.I.M.; Vongkamjan, K. Lysis profiles of Salmonella phages on Salmonella isolates from various sources and efficiency of a phage cocktail against S. Enteritidis and S. Typhimurium. Microorganisms 2019, 7, 100. [Google Scholar] [CrossRef]
- Sripaurya, B.; Ngasaman, R.; Benjakul, S.; Vongkamjan, K. Virulence genes and antibiotic resistance of Salmonella recovered from a wet market in Thailand. J. Food Saf. 2019, 39, e12601. [Google Scholar] [CrossRef]
- Pelyuntha, W.; Ngasaman, R.; Yingkajorn, M.; Chukiatsiri, K.; Vongkamjan, K. Decontamination of major Salmonella serovars derived from poultry farms on eggs using Salmonella phage cocktail. Asia Pac. J. Sci. Technol. 2022, 27, APST-27-02-16. [Google Scholar]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Switt, A.I.M.; den Bakker, H.C.; Vongkamjan, K.; Hoelzer, K.; Warnick, L.D.; Cummings, K.J.; Wiedmann, M. Salmonella bacteriophage diversity reflects host diversity on dairy farms. Food Microbiol. 2013, 36, 275–285. [Google Scholar] [CrossRef]
- Dueñas, F.; Rivera, D.; Toledo, V.; Tardone, R.; Hervé-Claude, L.P.; Hamilton-West, C.; Switt, A.I.M. Characterization of Salmonella phages from dairy calves on farms with history of diarrhea. J. Dairy Sci. 2017, 100, 2196–2200. [Google Scholar] [CrossRef] [PubMed]
- Duc, H.M.; Son, H.M.; Honjoh, K.I.; Miyamoto, T. Isolation and application of bacteriophages to reduce Salmonella contamination in raw chicken meat. LWT 2018, 91, 353–360. [Google Scholar] [CrossRef]
- Shang, Y.; Sun, Q.; Chen, H.; Wu, Q.; Chen, M.; Yang, S.; Du, M.; Zha, F.; Ye, Q.; Zhang, J. Isolation and characterization of a novel Salmonella phage vB_SalP_TR2. Front. Microbiol. 2021, 12, 1452. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.L.; Brüssow, H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.; Kutter, E.; Karriker, L.; Stahl, C.; Wagstrom, E.; Anderson, R.C.; Genovese, K.; McRaynolds, J.; et al. Occurrence of Salmonella-specific bacteriophages in swine feces collected from commercial farms. Foodborne Pathog. Dis. 2010, 7, 851–856. [Google Scholar] [CrossRef]
- Goodridge, L.D. Designing phage therapeutics. Curr. Pharm. Biotechnol. 2010, 11, 15–27. [Google Scholar] [CrossRef]
- Hooton, S.P.; Atterbury, R.J.; Connerton, I.F. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int. J. Food Microbiol. 2011, 151, 157–163. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology: Phage cocktails. Adv. Appl. Microbiol. 2012, 78, 1–23. [Google Scholar]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Heyse, S.; Hanna, L.F.; Woolston, J.; Sulakvelidze, A.; Charbonneau, D. Bacteriophage cocktail for biocontrol of Salmonella in dried pet food. J. Food Prot. 2015, 78, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Henein, A.E.; Hanlon, G.W.; Cooper, C.J.; Denyer, S.P.; Maillard, J.Y. A partially purified Acinetobacter baumannii phage preparation exhibits no cytotoxicity in 3T3 mouse fibroblast cells. Front. Microbiol. 2016, 7, 1198. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.; Ferreira, R.; Costa, A.R.; Oliveira, H.; Azeredo, J. Efficacy and safety assessment of two enterococci phages in an in vitro biofilm wound model. Sci. Rep. 2019, 9, 6643. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, V. Feed enzymes: The science, practice, and metabolic realities. J. Appl. Poult. Res. 2013, 22, 628–636. [Google Scholar] [CrossRef]
- Bardina, C.; Spricigo, D.A.; Cortés, P.; Llagostera, M. Significance of the bacteriophage treatment schedule in reducing Salmonella colonization of poultry. Appl. Environ. Microbiol. 2012, 78, 6600–6607. [Google Scholar] [CrossRef]
| Salmonella Serovars | Time (h) | Bacterial Count (log CFU/mL) 1 | ||
|---|---|---|---|---|
| Control | Phage Cocktail at Low MOI (103) | Phage Cocktail at High MOI (104) | ||
| Enteritidis | 0 | 3.9 ± 0.0 a | 3.9 ± 0.0 | 3.8 ± 0.4 |
| 6 | 7.7 ± 0.2 b | ND | ND | |
| 12 | 9.3 ± 0.1 c | ND | ND | |
| 18 | 9.5 ± 0.2 c | ND | ND | |
| Typhimurium | 0 | 3.9 ± 0.1 a | 3.9 ± 0.2 | 4.1 ± 0.2 |
| 6 | 8.0 ± 0.0 b | ND | ND | |
| 12 | 8.3 ± 0.1 b | ND | ND | |
| 18 | 9.6 ± 0.1 c | ND | ND | |
| Salmonella Serovar | Time (h) | Bacterial Count (log CFU/mL) 1 | |||
|---|---|---|---|---|---|
| Control | KP001 | KP005 | WP110 | ||
| Enteritidis | 0 | 3.8 ± 0.0 a | 3.7 ± 0.0 a | 3.6 ± 0.1 a | 3.8 ± 0.2 a |
| 6 | 7.5 ± 0.2 b | 4.9 ± 0.2 b* | 5.3 ± 0.3 b* | 5.2 ± 0.1 b* | |
| 12 | 8.3 ± 0.0 c | 7.6 ± 0.0 c* | 6.5 ± 0.0 c* | 7.4 ± 0.3 c* | |
| 18 | 9.2 ± 0.1 d | 8.1 ± 0.1 d* | 7.4 ± 0.0 d* | 7.6 ± 0.4 c* | |
| Typhimurium | 0 | 3.7 ± 0.3 a | 3.9 ± 0.3 a | 3.4 ± 0.3 a | 3.5 ± 0.1 a |
| 6 | 7.9 ± 0.2 b | 5.2 ± 0.1 b* | 4.4 ± 0.1 b* | 5.0 ± 0.0 b* | |
| 12 | 8.4 ± 0.2 c | 5.5 ± 0.0 b* | 6.7 ± 0.0 c* | 5.2 ± 0.1 b* | |
| 18 | 9.3 ± 0.0 d | 5.9 ± 0.2 b* | 6.5 ± 0.5 c* | 6.9 ± 0.2 c* | |
| Salmonella | Treatment | Lysis Ability of Cocktail 1 (Phage Titer log PFU/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Passage | |||||||||||||
| 1 | 2 | 3 | |||||||||||
| 7 | 6 | 5 | 4 | 7 | 6 | 5 | 4 | 7 | 6 | 5 | 4 | ||
| S. Enteritidis | Control | +++ | ++ | ++ | + | ++ | + | + | - | ++ | ++ | - | - |
| Cocktail | ++ | ++ | + | + | ++ | ++ | + | - | ++ | ++ | - | - | |
| S. Typhimurium | Control | +++ | +++ | ++ | + | +++ | ++ | + | - | +++ | ++ | + | - |
| Cocktail | +++ | ++ | + | + | +++ | +++ | + | + | +++ | ++ | + | - | |
| Phage | Time (h) | Cell Viability (%) | ||||
|---|---|---|---|---|---|---|
| Phage Concentration (log PFU/mL) 1 | ||||||
| Control | 6.0 | 7.0 | 8.0 | 9.0 | ||
| vB_SenS_KP001 | 24 | 100 | >100 | >100 | >100 | >100 |
| 48 | 100 | >100 | >100 | >100 | >100 | |
| 72 | 100 | >100 | >100 | >100 | >100 | |
| vB_SenS_KP005 | 24 | 100 | >100 | >100 | >100 | >100 |
| 48 | 100 | >100 | 92.4 ± 7.8 * | 95.4 ± 3.4 * | >100 | |
| 72 | 100 | >100 | >100 | >100 | >100 | |
| vB_SenS_WP110 | 24 | 100 | >100 | >100 | >100 | >100 |
| 48 | 100 | >100 | >100 | >100 | >100 | |
| 72 | 100 | >100 | >100 | >100 | >100 | |
| Phage | Time (h) | Cell Viability (%) 1 | ||||
|---|---|---|---|---|---|---|
| Phage Concentration (log PFU/mL) | ||||||
| Control | 6.0 | 7.0 | 8.0 | 9.0 | ||
| vB_SenS_KP001 | 24 | 100 | 96.2 ± 4.2 | 97.1 ± 3.5 | 96.1 ± 4.4 | 98.9 ± 3.7 |
| 48 | 100 | >100 | 99.5 ± 1.7 | 98.3 ± 2.3 | 98.6 ± 4.4 | |
| 72 | 100 | >100 | >100 | 95.9 ± 3.1 * | 97.6 ± 2.9 * | |
| vB_SenS_KP005 | 24 | 100 | 96.7 ± 2.2 | 96.3 ± 1.7 | 97.0 ± 3.7 | 96.0 ± 3.8 |
| 48 | 100 | >100 | 99.2 ± 2.1 | 98.6 ± 1.6 | 96.3 ± 5.6 | |
| 72 | 100 | >100 | >100 | 99.3 ± 1.5 | 96.9 ± 2.3 * | |
| vB_SenS_WP110 | 24 | 100 | 96.0 ± 2.8 * | 95.9 ± 2.9 * | 97.0 ± 2.7 | 99.1 ± 6.5 |
| 48 | 100 | 97.7 ± 2.7 * | 99.3 ± 1.9 | 99.8 ± 1.3 | 99.2 ± 2.0 | |
| 72 | 100 | 96.4 ± 1.2 * | 95.7 ± 3.3 * | 95.3 ± 4.2 * | 98.8 ± 3.1 | |
| Time of Treatment (Days) | Age of Broiler (Days) | Salmonella Prevalence (Positive Samples/Total Samples) (%) | ||
|---|---|---|---|---|
| Control | Feeding Tube | Nipple Waterer | ||
| 0 | 11 | 8/10 (80) | 9/10 (90) | 7/10 (70) |
| 4 | 15 | 7/10 (70) | 0/10 (0) | 4/10 (40) |
| 13 | 24 | 6/10 (60) | 0/10 (0) | 0/10 (0) |
| 16 | 27 | 10/10 (100) | 0/10(0) | 0/10 (0) |
| 20 | 31 | 10/10 (100) | 0/10 (0) | 0/10 (0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pelyuntha, W.; Yafa, A.; Ngasaman, R.; Yingkajorn, M.; Chukiatsiri, K.; Champoochana, N.; Vongkamjan, K. Oral Administration of a Phage Cocktail to Reduce Salmonella Colonization in Broiler Gastrointestinal Tract—A Pilot Study. Animals 2022, 12, 3087. https://doi.org/10.3390/ani12223087
Pelyuntha W, Yafa A, Ngasaman R, Yingkajorn M, Chukiatsiri K, Champoochana N, Vongkamjan K. Oral Administration of a Phage Cocktail to Reduce Salmonella Colonization in Broiler Gastrointestinal Tract—A Pilot Study. Animals. 2022; 12(22):3087. https://doi.org/10.3390/ani12223087
Chicago/Turabian StylePelyuntha, Wattana, Ananya Yafa, Ruttayaporn Ngasaman, Mingkwan Yingkajorn, Kridda Chukiatsiri, Nidanut Champoochana, and Kitiya Vongkamjan. 2022. "Oral Administration of a Phage Cocktail to Reduce Salmonella Colonization in Broiler Gastrointestinal Tract—A Pilot Study" Animals 12, no. 22: 3087. https://doi.org/10.3390/ani12223087
APA StylePelyuntha, W., Yafa, A., Ngasaman, R., Yingkajorn, M., Chukiatsiri, K., Champoochana, N., & Vongkamjan, K. (2022). Oral Administration of a Phage Cocktail to Reduce Salmonella Colonization in Broiler Gastrointestinal Tract—A Pilot Study. Animals, 12(22), 3087. https://doi.org/10.3390/ani12223087






