The Complete Mitochondrial Genome of Eurasian Minnow (Phoxinus cf. Phoxinus) from the Heilongjiang River, and Its Phylogenetic Implications
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Sequencing and Annotation of the Mitogenome
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Characterization of the Mitogenome
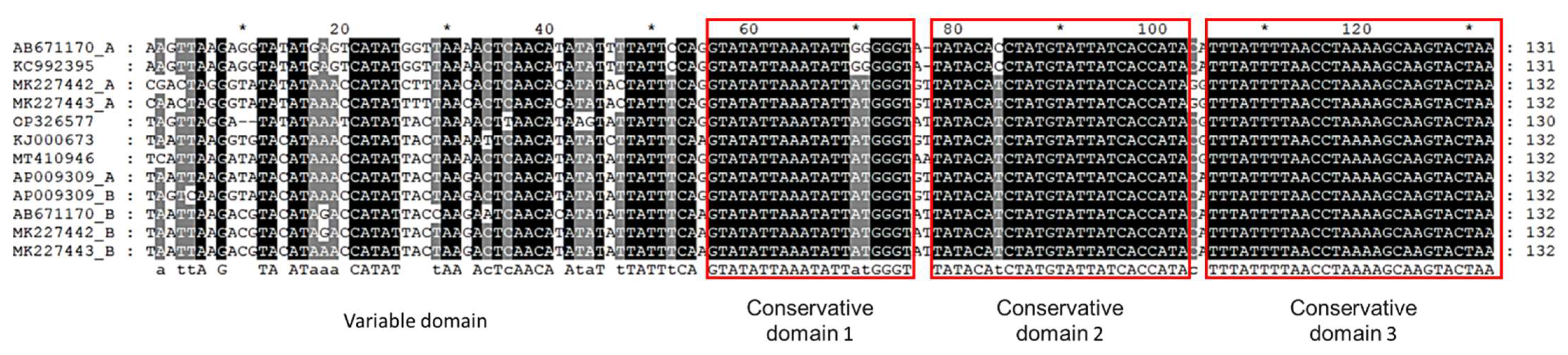
3.2. The D-loop Region of Phoxinus Spp.
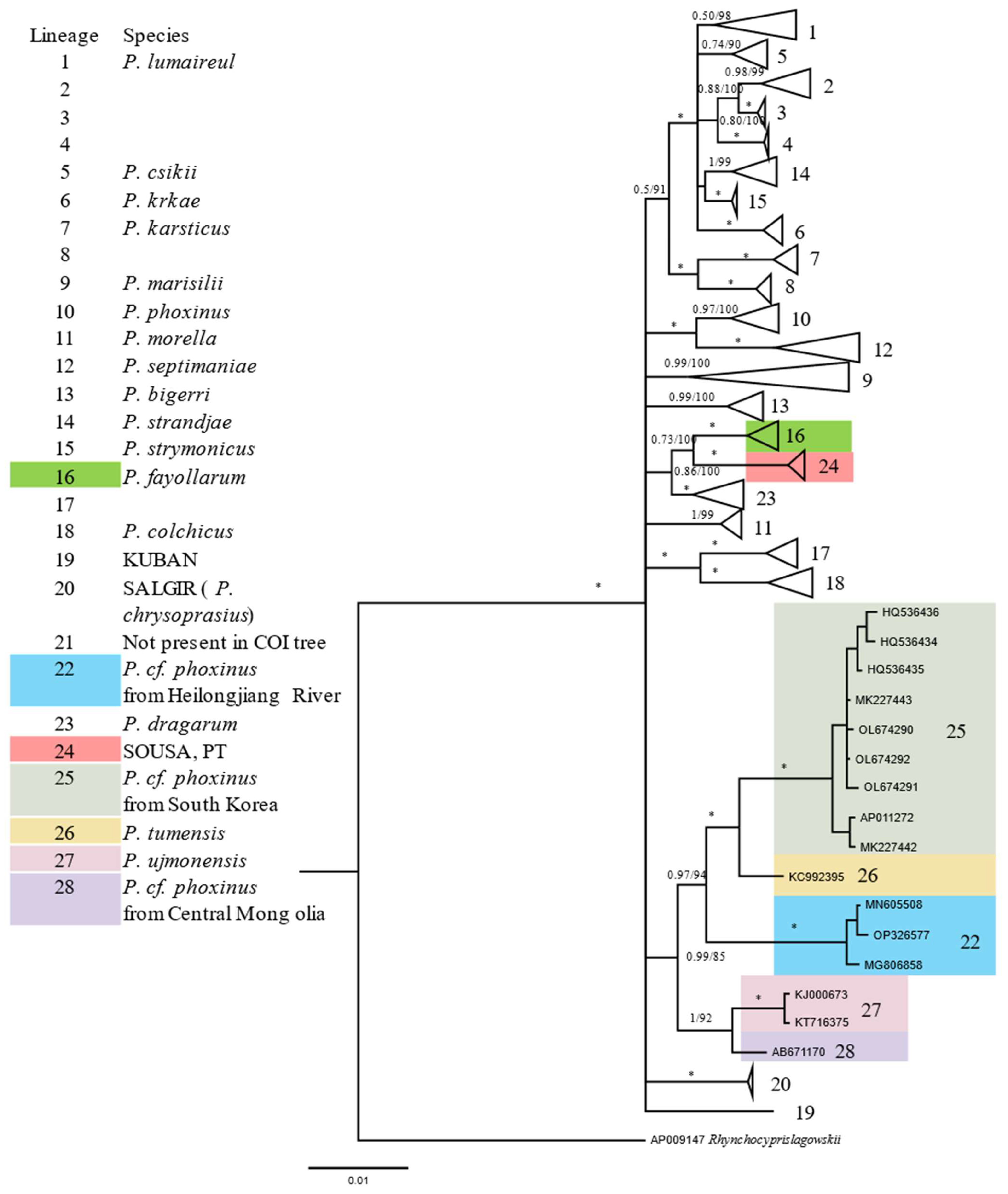
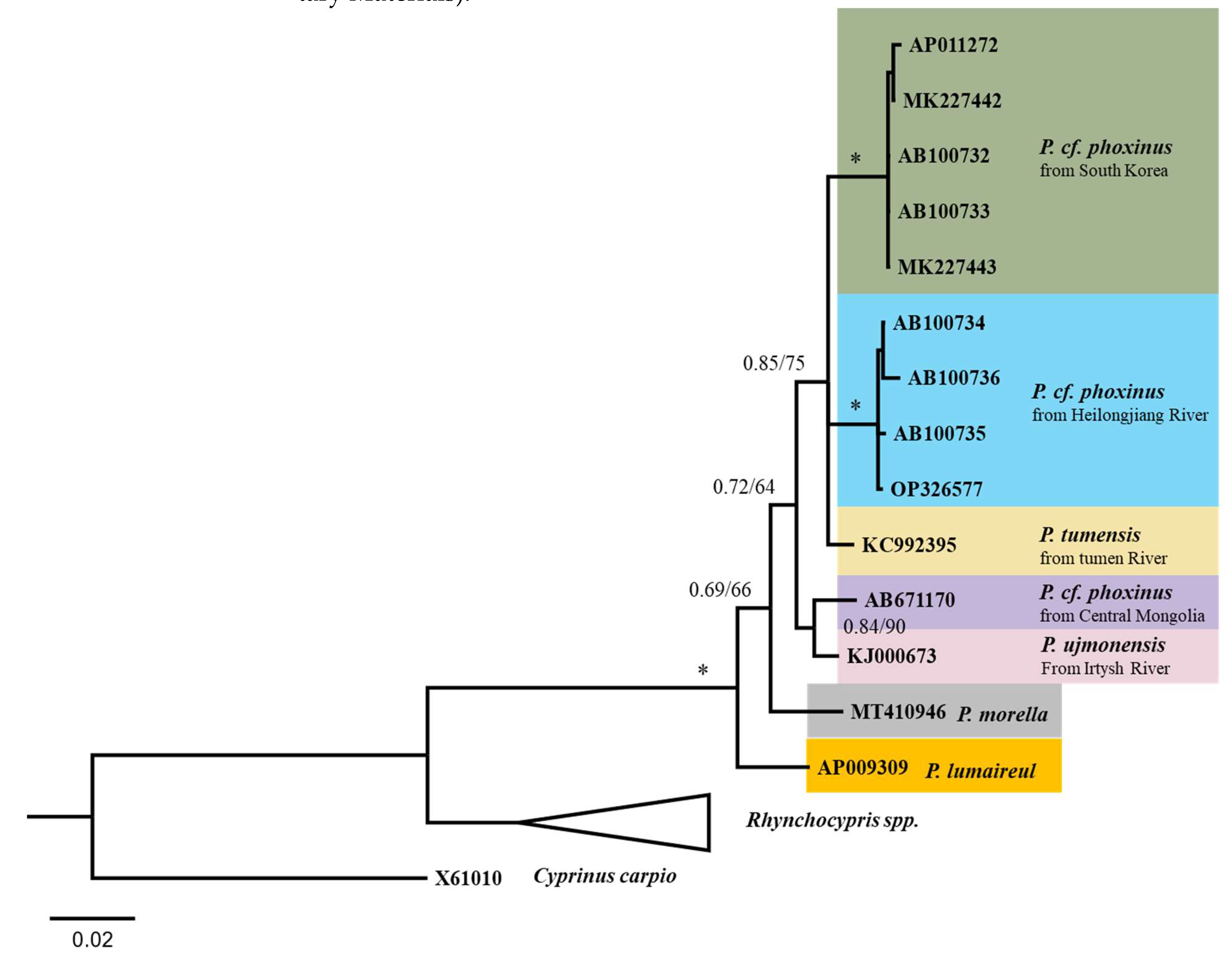
3.3. Phylogeny of Phoxinus Spp.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kottelat, M. Fishes of Mongolia. In A Check-List of the Fishes Known to Occur in Mongolia with Comments on Systematics and Nomenclature; The World Bank: Washington, DC, USA, 2006. [Google Scholar]
- Strange, R.M.; Mayden, R.L. Phylogenetic Relationships and a Revised Taxonomy for North American Cyprinids Currently Assigned to Phoxinus (Actinopterygii: Cyprinidae). Copeia 2009, 2009, 494–501. [Google Scholar] [CrossRef]
- Kottelat, M. Three new species of Phoxinus from Greece and southern France (Teleostei: Cyprinidae). Ichthyol. Explor. Fres. 2007, 18, 145–162. [Google Scholar]
- Bianco, P.G.; De Bonis, S. A Taxonomic Study on the Genus Phoxinus (Acthinopterigy, Cyprinidae) from Italy and Western Balkans with Description of Four New Species: P. ketmaieri, P. karsticus, P. apollonicus and P. likai. In Researches on Wildlife Conservation; Bianco, P.G., de Filippo, G., Eds.; IGF Publishing: Eugene, OR, USA, 2015; Volume 4. [Google Scholar]
- Palandačić, A.; Bravničar, J.; Zupančič, P.; Šanda, R.; Snoj, A. Molecular Data Suggest a Multispecies Complex of Phoxinus (Cyprinidae) in the Western Balkan Peninsula. Mol. Phylogenet. Evol. 2015, 92, 118–123. [Google Scholar] [CrossRef]
- Denys, G.P.J.; Dettai, A.; Persat, H.; Daszkiewicz, P.; Hautecoeur, M.; Keith, P. Revision of Phoxinus in France with the Description of Two New Species (Teleostei, Leuciscidae). Cybium 2020, 44, 205–237. [Google Scholar] [CrossRef]
- Palandačić, A.; Naseka, A.; Ramler, D.; Ahnelt, H. Contrasting Morphology with Molecular Data: An Approach to Revision of Species Complexes Based on the Example of European Phoxinus (Cyprinidae). BMC Evol. Biol. 2017, 17, 184. [Google Scholar] [CrossRef]
- Ito, Y.; Sakai, H.; Shedko, S.; Jeon, S.R. Genetic Differentiation of the Northern Far East Cyprinids, Phoxinus and Rhynchocypris. Fish. Sci. 2002, 68, 75–78. [Google Scholar] [CrossRef]
- Sakai, H.; Ito, Y.; Shedko, S.V.; Safronov, S.N.; Frolov, S.V.; Chereshnev, I.A.; Jeon, S.R.; Goto, A. Phylogenetic and Taxonomic Relationships of Northern Far Eastern Phoxinin Minnows, Phoxinus and Rhynchocypris (Pisces, Cyprinidae), as Inferred from Allozyme and Mitochondrial 16S RRNA Sequence Analyses. Zool. Sci. 2006, 23, 323–331. [Google Scholar] [CrossRef]
- Collin, H.; Fumagalli, L. Evidence for Morphological and Adaptive Genetic Divergence between Lake and Stream Habitats in European Minnows (Phoxinus phoxinus, Cyprinidae): Adaptive Divergence in European Minnow. Mol. Ecol. 2011, 20, 4490–4502. [Google Scholar] [CrossRef]
- Paśko, Ł.; Kusznierz, J.; Maślak, R.; Tagayev, D.; Sergiel, A.; Pietras-Lebioda, A.; Borczyk, B. Morphometric Exploration of Diversity of the Eurasian Minnow Phoxinus phoxinus: A Case Study of a Widely Distributed Palaearctic Fish. Ann. Zool. Fenn. 2014, 51, 399–412. [Google Scholar] [CrossRef]
- Collin, H.; Fumagalli, L. The Role of Geography and Ecology in Shaping Repeated Patterns of Morphological and Genetic Differentiation between European Minnows (Phoxinus phoxinus) from the Pyrenees and the Alps. Biol. J. Linn. Soc. 2015, 116, 691–703. [Google Scholar] [CrossRef]
- Ramler, D.; Palandačić, A.; Delmastro, G.B.; Wanzenböck, J.; Ahnelt, H. Morphological Divergence of Lake and Stream Phoxinus of Northern Italy and the Danube Basin Based on Geometric Morphometric Analysis. Ecol. Evol. 2017, 7, 572–584. [Google Scholar] [CrossRef]
- Scharnweber, K. Morphological and Trophic Divergence of Lake and Stream Minnows (Phoxinus phoxinus). Ecol. Evol. 2020, 10, 8358–8367. [Google Scholar] [CrossRef]
- Schönhuth, S.; Shiozawa, D.K.; Dowling, T.E.; Mayden, R.L. Molecular Systematics of Western North American Cyprinids (Cypriniformes: Cyprinidae). Zootaxa 2012, 3586, 281. [Google Scholar] [CrossRef]
- Corral-Lou, A.; Perea, S.; Aparicio, E.; Doadrio, I. Phylogeography and Species Delineation of the Genus Phoxinus Rafinesque, 1820 (Actinopterygii: Leuciscidae) in the Iberian Peninsula. J. Zool. Syst. Evol. Res. 2019, 57, 926–941. [Google Scholar] [CrossRef]
- Palandačić, A.; Kruckenhauser, L.; Ahnelt, H.; Mikschi, E. European Minnows through Time: Museum Collections Aid Genetic Assessment of Species Introductions in Freshwater Fishes (Cyprinidae: Phoxinus Species Complex). Heredity 2020, 124, 410–422. [Google Scholar] [CrossRef]
- Palandačić, A.; Witman, K.; Spikmans, F. Molecular Analysis Reveals Multiple Native and Alien Phoxinus Species (Leusciscidae) in the Netherlands and Belgium. Biol. Invasions. 2022, 24, 2273–2283. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Xiong, J.; Huang, Q.; Chen, M.; Fang, J. Identification of Species and Authenticity of Shark Fins by DNA Barcoding Technology. Chin. J. Fish. 2020, 33, 28–33. (In Chinese) [Google Scholar]
- Garcia-Raventós, A.; Martins, F.M.S.; Teixeira, A.; Sousa, R.; Froufe, E.; Varandas, S.; Lopes-Lima, M.; Beja, P.; Filipe, A.F. Origin and History of Phoxinus (Cyprinidae) Introductions in the Douro Basin (Iberian Peninsula): An Update Inferred from Genetic Data. Biol. Invasions. 2020, 22, 2409–2419. [Google Scholar] [CrossRef]
- De Santis, V.; Delmastro, G.B.; Vanetti, I.; Britton, J.R.; Zaccara, S. Species Composition of Introduced and Natural Minnow Populations of the Phoxinus Cryptic Complex in the Westernmost Part of the Po River Basin (North Italy). Biol. Invasions. 2021, 23, 657–668. [Google Scholar] [CrossRef]
- Xie, P.; Zhao, G.; Niu, J.; Wang, J.; Zhou, Q.; Guo, Y.; Ma, X. Comprehensive Analysis of Population Genetics of Phoxinus phoxinus ujmonensis in the Irtysh River: Abiotic and Biotic Factors. Ecol. Evol. 2019, 9, 7997–8012. [Google Scholar] [CrossRef]
- Yang, T.; Meng, W.; Zhang, R.; Gao, T.; Cai, L.; Hai, S.; Zhou, Q. DNA barcoding of fishes in Irtysh River China. Russ. J. Genet 2016, 52, 969–976. [Google Scholar] [CrossRef]
- Kartavtsev, Y.P.; Batischeva, N.M.; Bogutskaya, N.G.; Katugina, A.O.; Hanzawa, N. Molecular systematics and DNA barcoding of Altai osmans, oreoleuciscus (pisces, cyprinidae, and leuciscinae), and their nearest relatives, inferred from sequences of cytochrome b (Cyt-b), cytochrome oxidase c (Co-1), and complete mitochondrial genome. Mitochondrial DNA Part A 2017, 28, 502–517. [Google Scholar] [CrossRef]
- Kim, D.; Hirt, M.V.; Won, Y.-J.; Simons, A.M. Small fishes crossed a large mountain range: Quaternary stream capture events and freshwater fishes on both sides of the Taebaek Mountains. Integr. Zool. 2017, 12, 292–302. [Google Scholar] [CrossRef]
- Schönhuth, S.; Vukić, J.; Šanda, R.; Yang, L.; Mayden, R.L. Phylogenetic Relationships and Classification of the Holarctic Family Leuciscidae (Cypriniformes: Cyprinoidei). Mol. Phylogenet. Evol. 2018, 127, 781–799. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Klass, A.L.; Kondakov, A.V.; Vikhrev, I.V.; Bespalaya, Y.V.; Gofarov, M.Y. Freshwater mussels house a diverse mussel-associated leech assemblage. Sci. Rep. 2019, 9, 16449. [Google Scholar] [CrossRef]
- Lebedeva, D.I.; Chrisanfova, G.G.; Ieshko, E.P.; Guliaev, A.S.; Yakovleva, G.A.; Mendsaikhan, B.; Semyenova, S.K. Morphological and molecular differentiation of Diplostomum spp. metacercariae from brain of minnows (Phoxinus phoxinus L.) in four populations of northern Europe and East Asia. Infect. Genet. Evol. 2021, 92, 104911. [Google Scholar] [CrossRef]
- Miya, M.; Nishida, M. The Mitogenomic Contributions to Molecular Phylogenetetics and Evolution of Fishes: A 15-Year Retrospect. Ichthyol Res 2015, 62, 29–71. [Google Scholar] [CrossRef]
- Imoto, J.M.; Saitoh, K.; Sasaki, T.; Yonezawa, T.; Adachi, J.; Kartavtsev, Y.P.; Miya, M.; Nishida, M.; Hanzawa, N. Phylogeny and Biogeography of Highly Diverged Freshwater Fish Species (Leuciscinae, Cyprinidae, Teleostei) Inferred from Mitochondrial Genome Analysis. Gene 2013, 514, 112–124. [Google Scholar] [CrossRef]
- Xu, W.; Chen, A.; Xia, R.; Fu, C. Complete Mitochondrial Genome of Phoxinus Tumensis (Cypriniformes: Cyprinidae). Mitochondrial DNA 2014, 25, 368–369. [Google Scholar] [CrossRef]
- Xie, P.; Ao, M.; Liu, C.; Zhang, Z.; Zhang, Y.; Niu, J.; Karjan, A.; Ma, X. The Complete Mitochondrial Genome of Phoxinus Phoxinus Ujmonensis (Cypriniformes: Cyprinidae). Mitochondrial DNA 2016, 27, 212–213. [Google Scholar] [CrossRef]
- Lee, Y.J.; Cha, S.H.; An, J.; Suk, H.Y. The Complete Mitochondrial Genome Information of Phoxinus Phoxinus (Cypriniformes: Cyprinidae) on the Korean Peninsula and the Phylogenetic Implication. Mitochondrial DNA Part B 2019, 4, 3844–3845. [Google Scholar] [CrossRef]
- Luo, Y.L. Leuciscinae. In Fauna Sinica Osteichthyes: Cypriniformes (II); Science Press: Beijing, China, 1998; pp. 61–111. (In Chinese) [Google Scholar]
- Chang, Y.; Huang, F.; Lo, T. The Complete Nucleotide Sequence and Gene Organization of Carp (Cyprinus Carpio) Mitochondrial Genome. J. Mol. Evol. 1994, 38, 138–155. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A Mitochondrial Genome Database of Fish with an Accurate and Automatic Annotation Pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. TRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Liu, H.; Tzeng, C.-S.; Teng, H.-Y. Sequence Variations in the Mitochondrial DNA Control Region and Their Implications for the Phylogeny of the Cypriniformes. Can. J. Zool. 2002, 80, 569–581. [Google Scholar] [CrossRef]
- Broughton, R.E.; Milam, J.E.; Roe, B.A. The Complete Sequence of the Zebrafish (Danio Rerio) Mitochondrial Genome and Evolutionary Patterns in Vertebrate Mitochondrial DNA. Genome Res. 2001, 11, 1958–1967. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2, Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004.
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Buroker, N.E.; Brown, J.R.; Gilbert, T.A.; O’Hara, P.J.; Beckenbach, A.T.; Thomas, W.K.; Smith, M.J. Length Heteroplasmy of Sturgeon Mitochondrial DNA: An Illegitimate Elongation Model. Genetics 1990, 124, 157–163. [Google Scholar] [CrossRef]
- Zardoya, R.; Villalta, M.; López-Pérez, M.J.; Garrido-Pertierra, A.; Montoya, J.; Bautista, J.M. Nucleotide sequence of the sheep mitochondrial DNA D-loop and its flanking tRNA genes. Curr. Genet. 1995, 28, 94–96. [Google Scholar] [CrossRef]
- Madsen, C.S.; Ghivizzani, S.C.; Hauswirth, W.W. Protein Binding to a Single Termination-Associated Sequence in the Mitochondrial DNA D-Loop Region. Mol. Cell Biol. 1993, 13, 2162–2171. [Google Scholar] [CrossRef][Green Version]


| Species/Lineages | Mitogenomes | COI | Cyt b | 16s rRNA | Sample Size | Reference |
|---|---|---|---|---|---|---|
| P. cf. phoxinus | OP326577 | 1 | this study | |||
| (Heilongjiang River) | MG806858 | MG806682 | 1 | [27] | ||
| MN605508 | 1 | [28] | ||||
| AB100734-AB100736 | 44 | [9] | ||||
| P. tumensis | KC992395 | 1 | [32] | |||
| P. ujmonensis | KJ000673 | 1 | [33] | |||
| KR819892-KR819906 | 15 | [25] | ||||
| KT716375 | 1 | [24] | ||||
| P. cf. phoxinus | AB671170 | 1 | [31] | |||
| (Central Mongolia) | MW001939-MW001940 | 15 | [29] | |||
| P. cf. phoxinus | AP011272 | 1 | [31] | |||
| (South Korea) | MK227442 | 1 | [34] | |||
| MK227443 | 1 | [34] | ||||
| KX265376-KX265402 | 56 | [26] | ||||
| AB100732-AB100733 | 40 | [9] | ||||
| P. lumaireul | AP009309 | 1 | [31] | |||
| P. morella1 | MT410946 | N/A | Unpublished | |||
| Rhynchocypris lagowskii | AP009147 | 1 | [31] | |||
| Rhynchocypris spp. | AB100697-AB100731 | 582 | [9] | |||
| Cyprinus carpio | X61010 | X61010 | 1 | [36] |
| Gene/Element | From | to | Length(bp) | Start | Stop | Intergenic Nucleotides | Coding Strand |
|---|---|---|---|---|---|---|---|
| tRNA-Phe | 1 | 69 | 69 | ||||
| 12S rRNA | 70 | 1026 | 957 | ||||
| tRNA-Val | 1027 | 1098 | 72 | ||||
| 16S rRNA | 1099 | 2790 | 1692 | ||||
| tRNA-Leu (UAA) | 2791 | 2866 | 76 | ||||
| ND1 | 2868 | 3842 | 975 | ATG | TAG | 1 | |
| tRNA-Ile | 3847 | 3918 | 72 | 4 | |||
| tRNA-Gln | 3917 | 3987 | 71 | −2 | L | ||
| tRNA-Met | 3989 | 4057 | 69 | 1 | |||
| ND2 | 4058 | 5104 | 1047 | ATG | TAG | ||
| tRNA-Trp | 5103 | 5173 | 71 | −2 | |||
| tRNA-Ala | 5175 | 5243 | 69 | 1 | L | ||
| tRNA-Asn | 5245 | 5317 | 73 | 1 | L | ||
| OL | 5318 | 5348 | 31 | ||||
| tRNA-Cys | 5349 | 5416 | 68 | L | |||
| tRNA-Tyr | 5418 | 5488 | 71 | 1 | L | ||
| COI | 5490 | 7040 | 1551 | GTG | TAA | 1 | |
| tRNA-Ser (UGA) | 7041 | 7111 | 71 | L | |||
| tRNA-Asp | 7114 | 7187 | 74 | 2 | |||
| COII | 7202 | 7892 | 691 | ATG | T-- | 14 | |
| tRNA-Lys | 7893 | 7968 | 76 | ||||
| ATPase 8 | 7970 | 8134 | 165 | ATG | TAG | 1 | |
| ATPase 6 | 8128 | 8811 | 684 | ATG | TAA | −7 | |
| COIII | 8811 | 9595 | 785 | ATG | TA- | −1 | |
| tRNA-Gly | 9595 | 9665 | 71 | −1 | |||
| ND3 | 9666 | 10016 | 351 | GTG | TAG | ||
| tRNA-Arg | 10015 | 10083 | 69 | −2 | |||
| ND4L | 10084 | 10380 | 297 | ATG | TAA | ||
| ND4 | 10374 | 11756 | 1383 | ATG | TAG | −7 | |
| tRNA-His | 11756 | 11824 | 69 | −1 | |||
| tRNA-Ser (GCU) | 11825 | 11893 | 69 | ||||
| tRNA-Leu (UAG) | 11895 | 11967 | 73 | 1 | |||
| ND5 | 11968 | 13803 | 1836 | ATG | TAA | ||
| ND6 | 13800 | 14321 | 522 | ATG | TAA | −4 | L |
| tRNA-Glu | 14322 | 14390 | 69 | L | |||
| Cyt b | 14396 | 15536 | 1141 | ATG | T-- | 5 | |
| tRNA-Thr | 15537 | 15608 | 72 | ||||
| tRNA-Pro | 15608 | 15677 | 70 | −1 | L | ||
| control region | 15678 | 16789 | 1112 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, L.; Wang, E.; Li, W.; Yu, X.; Liao, X. The Complete Mitochondrial Genome of Eurasian Minnow (Phoxinus cf. Phoxinus) from the Heilongjiang River, and Its Phylogenetic Implications. Animals 2022, 12, 2960. https://doi.org/10.3390/ani12212960
Cheng L, Wang E, Li W, Yu X, Liao X. The Complete Mitochondrial Genome of Eurasian Minnow (Phoxinus cf. Phoxinus) from the Heilongjiang River, and Its Phylogenetic Implications. Animals. 2022; 12(21):2960. https://doi.org/10.3390/ani12212960
Chicago/Turabian StyleCheng, Lei, Ezhou Wang, Weitao Li, Xiaoli Yu, and Xiaolin Liao. 2022. "The Complete Mitochondrial Genome of Eurasian Minnow (Phoxinus cf. Phoxinus) from the Heilongjiang River, and Its Phylogenetic Implications" Animals 12, no. 21: 2960. https://doi.org/10.3390/ani12212960
APA StyleCheng, L., Wang, E., Li, W., Yu, X., & Liao, X. (2022). The Complete Mitochondrial Genome of Eurasian Minnow (Phoxinus cf. Phoxinus) from the Heilongjiang River, and Its Phylogenetic Implications. Animals, 12(21), 2960. https://doi.org/10.3390/ani12212960





