Molecular Detection and Genotyping of Enterocytozoon bieneusi in Beef Cattle in Shanxi Province, North China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Sampling
2.2. DNA Extraction and PCR Amplification
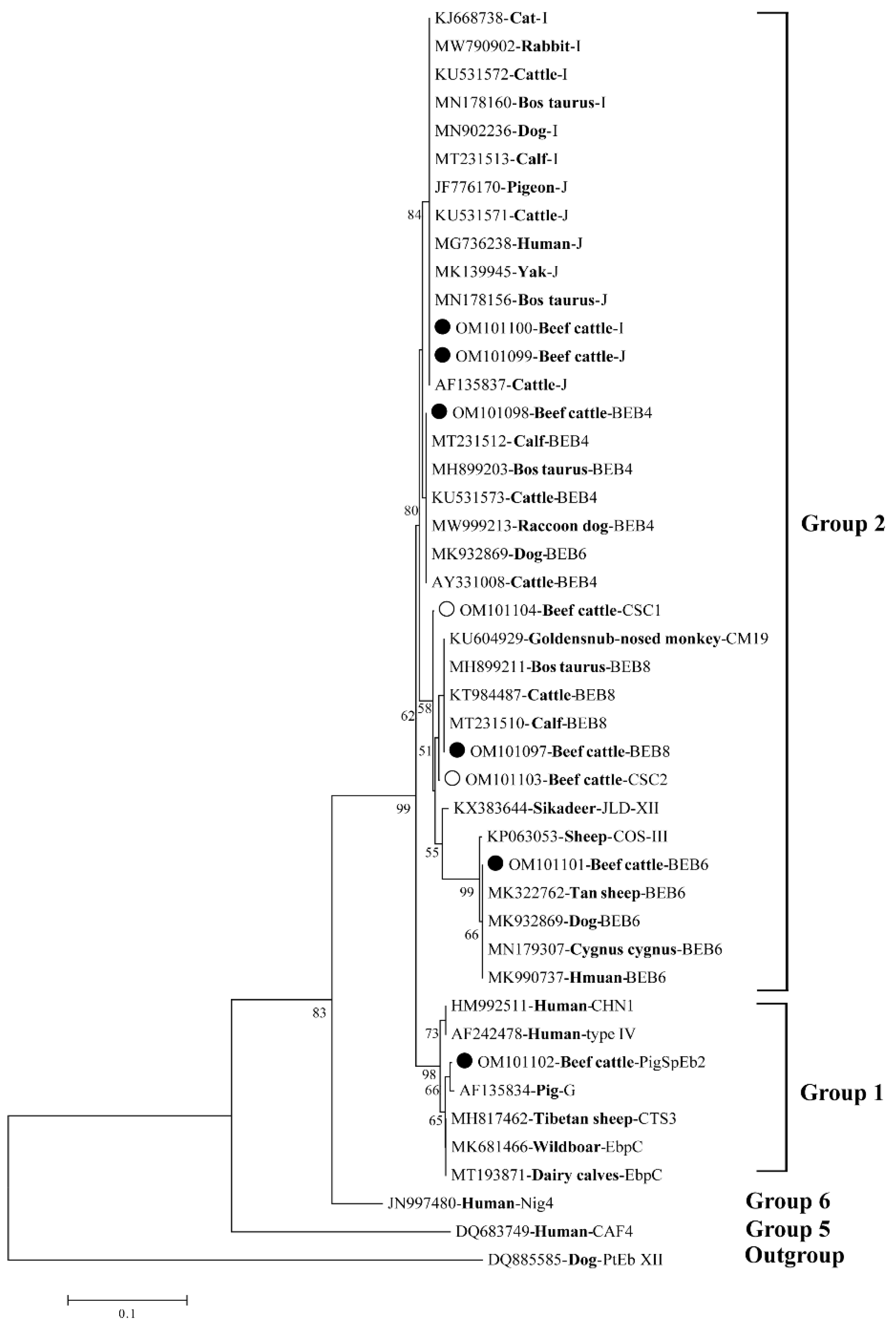
2.3. Sequence Analysis and Phylogenetic Reconstruction
2.4. Statistical Analysis
3. Results
3.1. Prevalence of E. bieneusi in Beef Cattle
3.2. E. bieneusi Genotype Contribution in Beef Cattle
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, B.; Pan, G.; Weiss, L.M. Microsporidiosis in humans. Clin. Microbiol. Rev. 2021, 34, e0001020. [Google Scholar] [CrossRef] [PubMed]
- Santin, M.; Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011, 90, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Didier, E.S.; Weiss, L.M. Microsporidiosis: Current status. Curr. Opin. Infect. Dis. 2006, 19, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Matos, O.; Lobo, M.L.; Xiao, L. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012, 2012, 981424. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Xia, W.; Li, W.; Ping, J.; Ding, S.; Liu, H. The prevalence of microsporidia in China: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 3174. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Zhang, L.; Xiao, L. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 2019, 75, 104033. [Google Scholar] [CrossRef]
- Li, W.; Feng, Y.; Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef]
- Rinder, H.; Thomschke, A.; Dengjel, B.; Gothe, R.; Loscher, T.; Zahler, M. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 2000, 86, 185–188. [Google Scholar]
- Fayer, R.; Santin, M.; Trout, J.M. Enterocytozoon bieneusi in mature dairy cattle on farms in the eastern United States. Parasitol. Res. 2007, 102, 15–20. [Google Scholar] [CrossRef]
- Zhang, Y.; Koehler, A.V.; Wang, T.; Haydon, S.R.; Gasser, R.B. Enterocytozoon bieneusi genotypes in cattle on farms located within a water catchment area. J. Eukaryot. Microbiol. 2019, 66, 553–559. [Google Scholar] [CrossRef]
- Abarca, N.; Santin, M.; Ortega, S.; Maloney, J.G.; George, N.S.; Molokin, A.; Cardona, G.A.; Dashti, A.; Koster, P.C.; Bailo, B.; et al. Molecular detection and characterization of Blastocystis sp. and Enterocytozoon bieneusi in cattle in northern Spain. Vet. Sci. 2021, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.; Rume, F.I.; Li, D.; Li, J.; Zhang, L. First molecular characterization of Enterocytozoon bieneusi in children and calves in Bangladesh. Transbound Emerg. Dis. 2022, 69, 1999–2007. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Luo, N.; Wang, C.; Qi, M.; Cao, J.; Cui, Z.; Huang, J.; Wang, R.; Zhang, L. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasites Vectors 2016, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H. Molecular detection of Enterocytozoon bieneusi and identification of a potentially human-pathogenic genotype in milk. Appl. Environ. Microbiol. 2008, 74, 1664–1666. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, Y.; Al, S.; Duzlu, O.; Onmaz, N.E.; Onder, Z.; Yetismis, G.; Hizlisoy, H.; Gonulalan, Z.; Yildirim, A. Enterocytozoon bieneusi in raw milk of cattle, sheep and water buffalo in Turkey: Genotype distributions and zoonotic concerns. Int. J. Food Microbiol. 2020, 334, 108828. [Google Scholar] [CrossRef]
- Li, N.; Xiao, L.; Wang, L.; Zhao, S.; Zhao, X.; Duan, L.; Guo, M.; Liu, L.; Feng, Y. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl. Trop. Dis. 2012, 6, e1809. [Google Scholar] [CrossRef]
- Taghipour, A.; Bahadory, S.; Abdoli, A. A systematic review and meta-analysis on the global prevalence of cattle microsporidiosis with focus on Enterocytozoon bieneusi: An emerging zoonotic pathogen. Prev. Vet. Med. 2022, 200, 105581. [Google Scholar] [CrossRef]
- Ruan, Y.; Xu, X.; He, Q.; Li, L.; Guo, J.; Bao, J.; Pan, G.; Li, T.; Zhou, Z. The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasites Vectors 2021, 14, 186. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Fayer, R.; Lal, A.A.; Trout, J.M.; Schaefer, F.W., 3rd; Xiao, L. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003, 69, 4495–4501. [Google Scholar] [CrossRef]
- Santin, M.; Fayer, R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: A consensus. J. Eukaryot. Microbiol. 2009, 56, 34–38. [Google Scholar] [CrossRef]
- Zhao, A.; Zhang, K.; Xu, C.; Wang, T.; Qi, M.; Li, J. Longitudinal identification of Enterocytozoon bieneusi in dairy calves on a farm in Southern Xinjiang, China. Comp. Immunol. Microbiol. Infect. Dis. 2020, 73, 101550. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Cai, M.; Wang, L.; Guo, Y.; Li, N.; Feng, Y.; Xiao, L. Genetic diversity within dominant Enterocytozoon bieneusi genotypes in pre-weaned calves. Parasites Vectors 2018, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, W.; Yang, F.; Zhang, L.; Wang, R.; Cao, J.; Shen, Y.; Liu, A. Enterocytozoon bieneusi in dairy cattle in the northeast of China: Genetic diversity of ITS gene and evaluation of zoonotic transmission potential. J. Eukaryot. Microbiol. 2015, 62, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santin, M.; Macarisin, D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol. Res. 2012, 111, 1349–1355. [Google Scholar] [CrossRef]
- Santin, M.; Dargatz, D.; Fayer, R. Prevalence and genotypes of Enterocytozoon bieneusi in weaned beef calves on cow-calf operations in the USA. Parasitol. Res. 2012, 110, 2033–2041. [Google Scholar] [CrossRef]
- Liu, X.; Tang, L.; Li, W.; Li, C.; Gu, Y. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J. Vet. Med. Sci. 2022, 84, 40–47. [Google Scholar] [CrossRef]
- Ma, J.G.; Zhang, N.Z.; Hou, J.L.; Zou, Y.; Hu, G.X.; Zhu, X.Q.; Zhou, D.H. Detection of Enterocytozoon bieneusi in white yaks in Gansu Province, China. Biomed. Res. Int. 2017, 2017, 5790181. [Google Scholar] [CrossRef]
- Feng, Y.; Gong, X.; Zhu, K.; Li, N.; Yu, Z.; Guo, Y.; Weng, Y.; Kvac, M.; Feng, Y.; Xiao, L. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasites Vectors 2019, 12, 41. [Google Scholar] [CrossRef]
- Xue, N.Y.; Liu, F.; Tao, W.F.; Zhao, Q.; Qiu, H.Y.; Hu, Y.; Chen, Y.; Wei, X.Y.; Wang, W.; Gao, D.; et al. Molecular detection of Cryptosporidium spp. and Enterocytozoon bieneusi in Longjiang Wagyu cattle in Northeastern China. Microb. Pathog. 2020, 149, 104526. [Google Scholar] [CrossRef]
- Zheng, X.L.; Zhou, H.H.; Ren, G.; Ma, T.M.; Cao, Z.X.; Wei, L.M.; Liu, Q.W.; Wang, F.; Zhang, Y.; Liu, H.L.; et al. Genotyping and zoonotic potential of Enterocytozoon bieneusi in cattle farmed in Hainan Province, the southernmost region of China. Parasite 2020, 27, 65. [Google Scholar] [CrossRef]
- Yu, F.; Qi, M.; Zhao, Z.; Lv, C.; Wang, Y.; Wang, R.; Zhang, L. The potential role of synanthropic rodents and flies in the transmission of Enterocytozoon bieneusi on a dairy cattle farm in China. J. Eukaryot. Microbiol. 2019, 66, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, P.; Zhu, X.Q.; Zou, Y.; Chen, X.Q. Prevalence and genotypes/subtypes of Enterocytozoon bieneusi and Blastocystis sp. in different breeds of cattle in Jiangxi Province, southeastern China. Infect. Genet. Evol. 2022, 98, 105216. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, N.; Jiang, W.; Guo, Y.; Wang, X.; Jin, Y.; Feng, Y.; Xiao, L. Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitol. Res. 2019, 118, 3053–3060. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Cai, J.; Ma, J.; Feng, Y.; Xiao, L. Enterocytozoon bieneusi genotypes in yaks (Bos grunniens) and their public health potential. J. Eukaryot. Microbiol. 2015, 62, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cai, J.; Li, P.; Wang, L.; Guo, Y.; Li, C.; Lei, M.; Feng, Y.; Xiao, L. Enterocytozoon bieneusi genotypes in Tibetan sheep and yaks. Parasitol. Res. 2018, 117, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Wang, R.J.; Ren, G.J.; Yu, Z.Q.; Zhang, L.X.; Zhang, S.Y.; Lu, H.; Peng, X.Q.; Zhao, G.H. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi Province, northwestern China. Parasitol. Res. 2016, 115, 1355–1361. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, Y.; Chang, Y.; Zhang, X.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Li, J.; et al. Genotyping and identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from free-range Tibetan yellow cattle and cattle-yak in Tibet, China. Acta Trop. 2020, 212, 105671. [Google Scholar] [CrossRef]
- Qi, M.; Jing, B.; Jian, F.; Wang, R.; Zhang, S.; Wang, H.; Ning, C.; Zhang, L. Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, Northwest China. Parasitol. Int. 2017, 66, 960–963. [Google Scholar] [CrossRef]
- Song, H.Y.; Wang, K.S.; Yang, J.F.; Mao, H.M.; Pu, L.H.; Zou, Y.; Ma, J.; Zhu, X.Q.; Zou, F.C.; He, J.J. Prevalence and novel genotypes identification of Enterocytozoon bieneusi in dairy cattle in Yunnan Province, China. Animals 2021, 11, 3014. [Google Scholar] [CrossRef]
- Thellier, M.; Breton, J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 2008, 15, 349–358. [Google Scholar] [CrossRef]
- Del Coco, V.F.; Cordoba, M.A.; Bilbao, G.; de Almeida Castro, P.; Basualdo, J.A.; Santin, M. First report of Enterocytozoon bieneusi from dairy cattle in Argentina. Vet. Parasitol. 2014, 199, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chang, Y.; Zhang, X.; Chen, Y.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Jian, F.; et al. Molecular characterization and distribution of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi from yaks in Tibet, China. BMC Vet. Res. 2019, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Dashti, A.; Rivero-Juarez, A.; Santin, M.; George, N.S.; Koster, P.C.; Lopez-Lopez, P.; Risalde, M.A.; Garcia-Bocanegra, I.; Gomez-Villamandos, J.C.; Caballero-Gomez, J.; et al. Diarrhoea-causing enteric protist species in intensively and extensively raised pigs (Sus scrofa domesticus) in Southern Spain. Part I: Prevalence and genetic diversity. Transbound. Emerg. Dis. 2021, 69, e1051–e1064. [Google Scholar] [CrossRef] [PubMed]
- Didier, E.S.; Weiss, L.M. Microsporidiosis: Not just in AIDS patients. Curr. Opin. Infect. Dis. 2011, 24, 490–495. [Google Scholar] [CrossRef]

| Factor | Categories | No. Tested | No. Positive | Prevalence% (95%CI) | OR (95%CI) | p-Value |
|---|---|---|---|---|---|---|
| Area | Qi | 177 | 35 | 19.77 (13.91–25.64) | 1 | 0.255 |
| Jishan | 224 | 55 | 24.55 (18.92–30.19) | 1.32 (0.82–2.13) | ||
| Gender | Male | 192 | 54 | 28.13 (21.77–34.48) | 1.88 (1.17–3.03) | <0.01 |
| Female | 209 | 36 | 17.22 (12.11–22.34) | 1 | ||
| Age | Age > 12 months | 258 | 49 | 18.99 (14.21–23.78) | 1 | <0.05 |
| Age ≤ 12 months | 143 | 41 | 28.67 (21.26–36.08) | 1.71 (1.06–2.76) | ||
| Total | 401 | 90 | 22.44 (18.36–26.53) |
| Factors | Categories | No. Positive/Total | Prevalence (%) | ITS Genotypes (n) |
|---|---|---|---|---|
| Region | Qi | 35/177 | 19.77 | I (15), J (6), BEB8 (4), BEB4(4), BEB6 (3), PigSpEb2 (1), CSC1 (1), CSC2 (1) |
| Jishan | 55/224 | 24.55 | I (35), BEB4 (16), J (4) | |
| Age | Age ≤ 12 months | 41/143 | 28.67 | I (18), BEB4 (11), J (5), BEB8 (3), BEB6 (3), PigSpEb2 (1) |
| Age > 12 months | 49/258 | 18.99 | I (32), BEB4 (9), J (5), BEB8 (1), CSC1 (1), CSC2 (1) | |
| Gender | Male | 54/192 | 28.13 | I (31), BEB4 (14), J (4), BEB8 (2), BEB6 (2), PigSpEb2 (1) |
| Female | 36/209 | 17.22 | I (19), J (6), BEB4 (6), BEB8 (2), BEB6 (1), CSC1 (1), CSC2 (1) | |
| Total | 90/401 | 22.44 | I (50), BEB4 (20), J (10), BEB8 (4), BEB6 (3), PigSpEb2 (1), CSC1 (1), CSC2 (1) |
| Province | Gene | No. Positive/Examined | Prevalence (%) | Prevalent Genotype | Host | References |
|---|---|---|---|---|---|---|
| Anhui | ITS | 40/955 | 4.19 | J | Cattle | [26] |
| Gansu | ITS | 4/353 | 1.13 | BEB4 | Yak | [27] |
| Guangdong | ITS | 61/388 | 15.72 | J | Pre-weaned cattle | [28] |
| Heilongjiang | ITS | 30/423 | 7.09 | J | Cattle | [29] |
| Hainan | ITS | 31/314 | 9.87 | EbpC | Cattle | [30] |
| Henan | ITS | 33/277 | 11.91 | BEB6 | Dairy cattle | [31] |
| Henan | ITS | 214/879 | 24.35 | J | Dairy cattle | [13] |
| Heilongjiang | ITS | 40/133 | 30.08 | O | Dairy cattle | [23] |
| Jiangxi | ITS | 30/556 | 5.40 | D | Cattle | [32] |
| Jiangsu | ITS | 177/1366 | 12.96 | J | Dairy cattle | [33] |
| Ningxia | ITS | 50/109 | 45.87 | J | Dairy cattle | [13] |
| Qinghai | ITS | 23/327 | 7.03 | BEB4 | Yak | [34] |
| Qinghai | ITS | 40/544 | 7.35 | J | Yak | [35] |
| Shanghai | ITS | 214/809 | 26.45 | J | Pre-weaned cattle | [22] |
| Shaanxi | ITS | 73/371 | 19.68 | I | Cattle | [36] |
| Tibet | ITS | 11/442 | 2.49 | I | Cattle | [37] |
| Xinjiang | ITS | 85/514 | 16.54 | J | Dairy cattle | [38] |
| Xinjiang | ITS | 130/250 | 52.00 | J | Dairy cattle | [21] |
| Yunnan | ITS | 5/841 | 0.59 | - a | Dairy cattle | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-Y.; Qin, R.-L.; Mei, J.-J.; Zou, Y.; Zhang, Z.-H.; Zheng, W.-B.; Liu, Q.; Zhu, X.-Q.; Gao, W.-W.; Xie, S.-C. Molecular Detection and Genotyping of Enterocytozoon bieneusi in Beef Cattle in Shanxi Province, North China. Animals 2022, 12, 2961. https://doi.org/10.3390/ani12212961
Liu Y-Y, Qin R-L, Mei J-J, Zou Y, Zhang Z-H, Zheng W-B, Liu Q, Zhu X-Q, Gao W-W, Xie S-C. Molecular Detection and Genotyping of Enterocytozoon bieneusi in Beef Cattle in Shanxi Province, North China. Animals. 2022; 12(21):2961. https://doi.org/10.3390/ani12212961
Chicago/Turabian StyleLiu, Ya-Ya, Rui-Lin Qin, Jin-Jin Mei, Yang Zou, Zhen-Huan Zhang, Wen-Bin Zheng, Qing Liu, Xing-Quan Zhu, Wen-Wei Gao, and Shi-Chen Xie. 2022. "Molecular Detection and Genotyping of Enterocytozoon bieneusi in Beef Cattle in Shanxi Province, North China" Animals 12, no. 21: 2961. https://doi.org/10.3390/ani12212961
APA StyleLiu, Y.-Y., Qin, R.-L., Mei, J.-J., Zou, Y., Zhang, Z.-H., Zheng, W.-B., Liu, Q., Zhu, X.-Q., Gao, W.-W., & Xie, S.-C. (2022). Molecular Detection and Genotyping of Enterocytozoon bieneusi in Beef Cattle in Shanxi Province, North China. Animals, 12(21), 2961. https://doi.org/10.3390/ani12212961





