chi-miR-99b-3p Regulates the Proliferation of Goat Skeletal Muscle Satellite Cells In Vitro by Targeting Caspase-3 and NCOR1
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cell Transfection
2.2. RNA Isolation and qRT-PCR
2.3. Western Blotting
2.4. Dual Luciferase Reporter Assay
2.5. Cell Proliferation Assays
2.6. Transcriptome Sequencing Analysis
2.7. Small RNA Sequencing Analysis
2.8. DEmiRNA and DEG Interaction Analysis
2.9. Statistical Analysis
3. Results
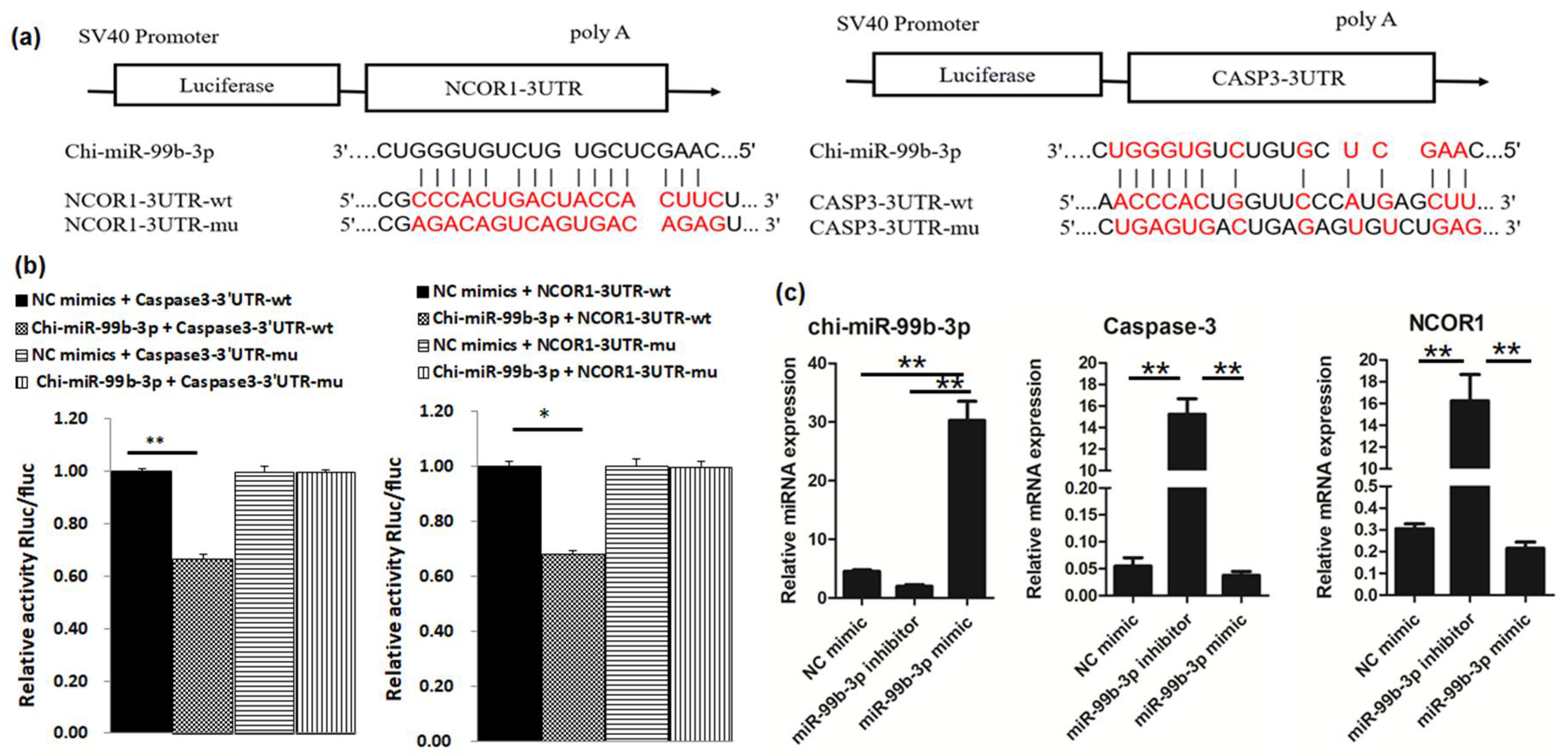
3.1. Caspase-3 and NCOR1 Are Targeted by chi-miR-99b-3p
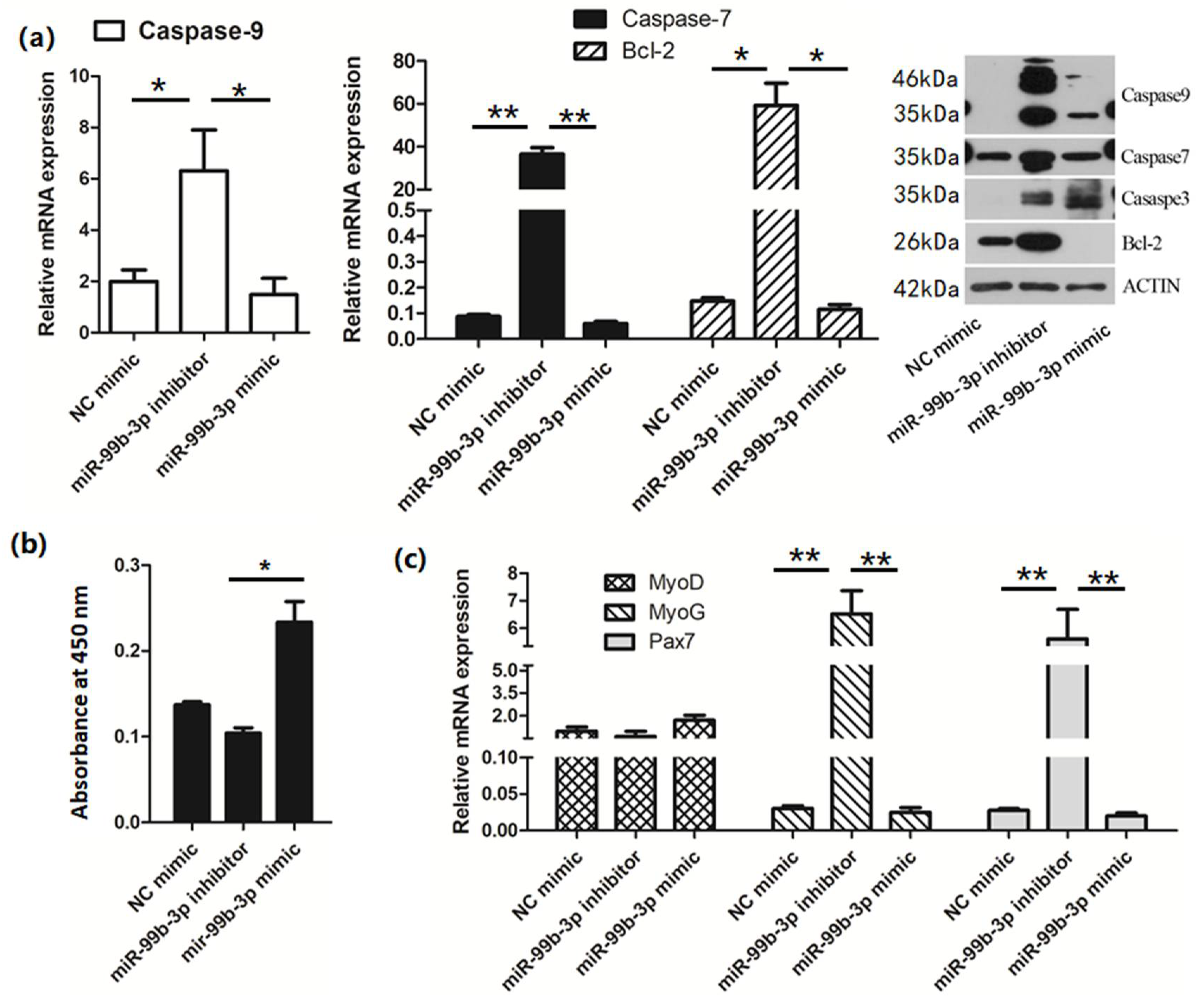
3.2. chi-miR-99b-3p Targets Caspase-3 and NCOR1 to Regulate Cell Proliferation and Differentiation
3.3. mRNA Profile Analysis after Upregulation of chi-miR-99b-3p
3.4. miRNA Profile Analysis after chi-miR-99b-3p Upregulation
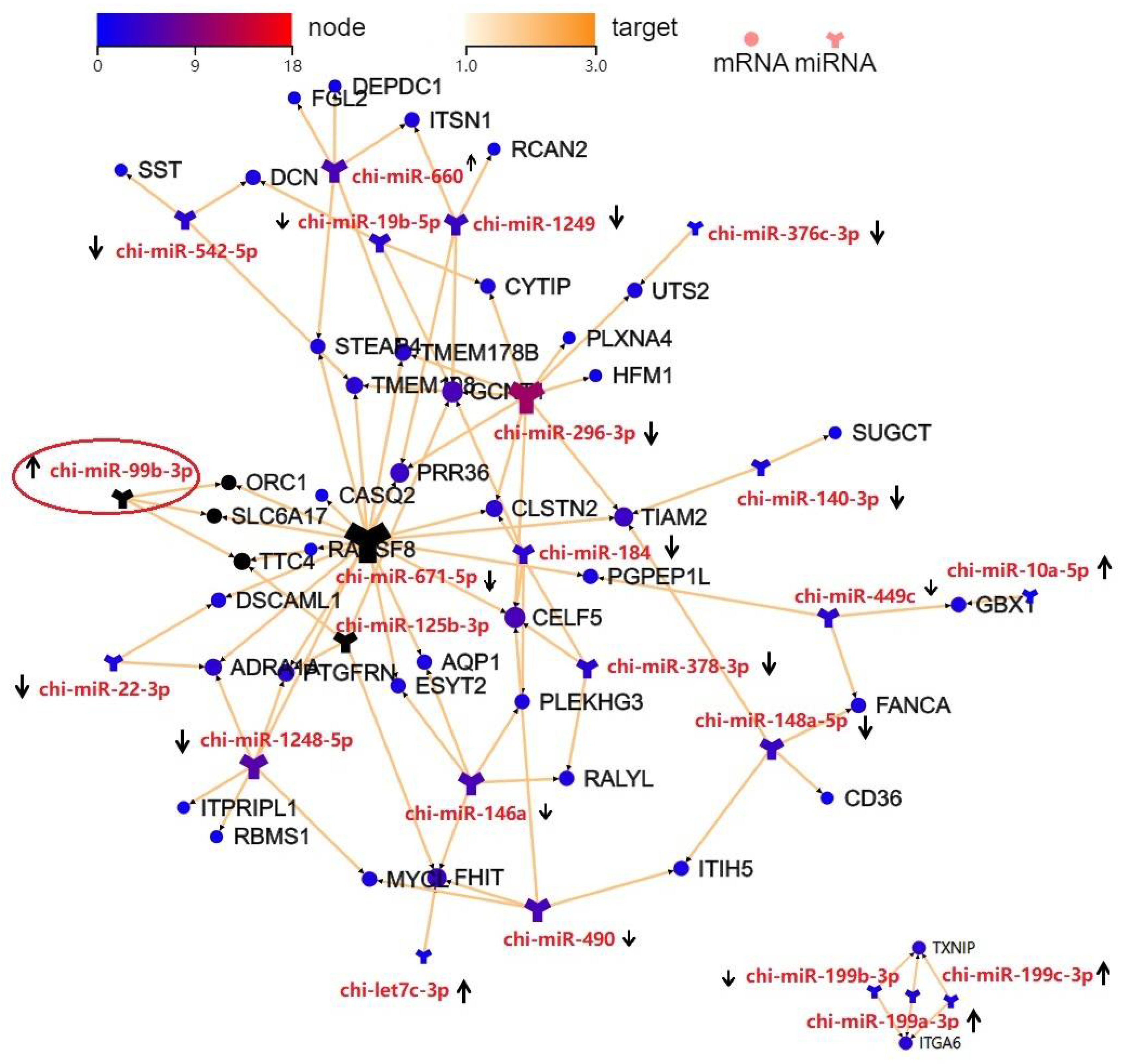
3.5. Integration Analysis of the DE miRNAs and DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mukund, K.; Subramaniam, S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1462. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, W.; Wang, S.; Ge, L.; Cao, X.; Wang, S.; Yuan, Z.; Lv, X.; Getachew, T.; Mwacharo, J.M.; et al. MicroRNA-181a regulates the proliferation and differentiation of Hu sheep skeletal muscle satellite cells and targets the YAP1 gene. Genes 2022, 13, 520. [Google Scholar] [CrossRef] [PubMed]
- Sui, M.; Zheng, Q.; Wu, H.; Zhu, L.; Ling, Y.; Wang, L.; Fang, F.; Liu, Y.; Zhang, Z.; Chu, M.; et al. The expression and regulation of miR-1 in goat skeletal muscle and satellite cell during muscle growth and development. Anim. Biotechnol. 2020, 31, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Lyu, M.; Wang, X.; Meng, X.; Qian, H.; Li, Q.; Ma, B.; Zhang, Z.; Xu, K. Chi-miR-487b-3p inhibits goat myoblast proliferation and differentiation by targeting IRS1 through the IRS1/PI3K/Akt signaling pathway. Int. J. Mol. Sci. 2021, 23, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, A.; Li, B.; Yuan, Y.; Lv, L.; Li, Y.; Wu, J.; Yang, L.; Bian, X.; Wang, K.; Wang, Q.; et al. Lactobacillus salivarius LI01 encapsulated in alginate-pectin microgels ameliorates D-galactosamine-induced acute liver injury in rats. Appl. Microbiol. Biotechnol. 2020, 104, 7437–7455. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Xu, H.; Lu, Y.; He, Q.; Yan, C.; Zhao, X.; Tian, Y.; Yang, C.; Zhang, Z.; Qiu, M.; et al. MUSTN1 is an indispensable factor in the proliferation, differentiation and apoptosis of skeletal muscle satellite cells in chicken. Exp. Cell Res. 2021, 407, 112833. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Liu, C.; Li, Y.; Gao, S.; Han, Y.C.; Wang, X.; Du, J.; Zhang, C. MicroRNA-223-3p promotes skeletal muscle regeneration by regulating inflammation in mice. J. Biol. Chem. 2020, 295, 10212–10223. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, H.; Liu, Y.; Liu, X.; Wang, X.; Sun, X.; Cheng, Y. MiR-99b-3p promotes hepatocellular carcinoma metastasis and proliferation by targeting protocadherin 19. Gene 2019, 698, 141–149. [Google Scholar] [CrossRef]
- Jakob, M.; Mattes, L.M.; Küffer, S.; Unger, K.; Hess, J.; Bertlich, M.; Haubner, F.; Ihler, F.; Canis, M.; Weiss, B.G.; et al. MicroRNA expression patterns in oral squamous cell carcinoma: Hsa-miR-99b-3p and hsa-miR-100-5p as novel prognostic markers for oral cancer. Head Neck 2019, 41, 3499–3515. [Google Scholar] [CrossRef]
- Chang, S.; Gao, Z.; Yang, Y.; He, K.; Wang, X.; Wang, L.; Gao, N.; Li, H.; He, X.; Huang, C. MiR-99b-3p is induced by vitamin D3 and contributes to its antiproliferative effects in gastric cancer cells by targeting HoxD3. Biol. Chem. 2019, 400, 1079–1086. [Google Scholar] [CrossRef]
- Yu, Y.H.; Zhang, Y.H.; Ding, Y.Q.; Bi, X.Y.; Yuan, J.; Zhou, H.; Wang, P.X.; Zhang, L.L.; Ye, J.T. MicroRNA-99b-3p promotes angiotensin II-induced cardiac fibrosis in mice by targeting GSK-3β. Acta Pharmacol. Sin. 2021, 42, 715–725. [Google Scholar] [CrossRef]
- Mitchell, C.J.; D’Souza, R.F.; Schierding, W.; Zeng, N.; Ramzan, F.; O’Sullivan, J.M.; Poppitt, S.D.; Cameron-Smith, D. Identification of human skeletal muscle miRNA related to strength by high-throughput sequencing. Physiol. Genom. 2018, 50, 416–424. [Google Scholar] [CrossRef]
- Liao, R.; Lv, Y.; Zhu, L.; Lin, Y. Altered expression of miRNAs and mRNAs reveals the potential regulatory role of miRNAs in the developmental process of early weaned goats. PLoS ONE 2019, 14, e0220907. [Google Scholar] [CrossRef]
- Wang, S.; Ma, T.; Zhao, G.; Zhang, N.; Tu, Y.; Li, F.; Cui, K.; Bi, Y.; Ding, H.; Diao, Q. Effect of age and weaning on growth performance, rumen fermentation, and serum parameters in lambs fed starter with limited ewe-lamb interaction. Animals 2019, 9, 825. [Google Scholar] [CrossRef]
- Asakura, A.; Hirai, H.; Kablar, B.; Morita, S.; Ishibashi, J.; Piras, B.A.; Christ, A.J.; Verma, M.; Vineretsky, K.A.; Rudnicki, M.A. Increased survival of muscle stem cells lacking the MyoD gene after transplantation into regenerating skeletal muscle. Proc. Natl. Acad. Sci. USA 2007, 104, 16552–16557. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, J.; Liu, W.; Wang, H.; Zhao, L.; Liu, S.; Li, P.; Zhang, S.; Sun, C.; Wu, Y.; et al. MicroRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, E10849–E10858. [Google Scholar] [CrossRef]
- Lu, Z.; Du, L.; Liu, R.; Di, R.; Zhang, L.; Ma, Y.; Li, Q.; Liu, E.; Chu, M.; Wei, C. MiR-378 and BMP-Smad can influence the proliferation of sheep myoblast. Gene 2018, 674, 143–150. [Google Scholar] [CrossRef]
- Kowalski, K.; Kołodziejczyk, A.; Sikorska, M.; Płaczkiewicz, J.; Cichosz, P.; Kowalewska, M.; Stremińska, W.; Jańczyk-Ilach, K.; Koblowska, M.; Fogtman, A.; et al. Stem cells migration during skeletal muscle regeneration-the role of Sdf-1/Cxcr4 and Sdf-1/Cxcr7 axis. Cell Adhes. Migr. 2017, 11, 384–398. [Google Scholar] [CrossRef]
- Zanou, N.; Gailly, P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell. Mol. Life Sci. 2013, 70, 4117–4130. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, G.; Slack, J.M. Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development 2006, 133, 2303–2313. [Google Scholar] [CrossRef] [Green Version]
- Budihardjo, I.; Oliver, H.; Lutter, M.; Luo, X.; Wang, X. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 1999, 15, 269–290. [Google Scholar] [CrossRef]
- Boivin, M.A.; Battah, S.I.; Dominic, E.A.; Kalantar-Zadeh, K.; Ferrando, A.; Tzamaloukas, A.H.; Dwivedi, R.; Ma, T.A.; Moseley, P.; Raj, D.S. Activation of caspase-3 in the skeletal muscle during haemodialysis. Eur. J. Clin. Investig. 2010, 40, 903–910. [Google Scholar] [CrossRef]
- Plant, P.J.; Bain, J.R.; Correa, J.E.; Woo, M.; Batt, J. Absence of caspase-3 protects against denervation-induced skeletal muscle atrophy. J. Appl. Physiol. (1985) 2009, 107, 224–234. [Google Scholar] [CrossRef]
- Yamamoto, H.; Williams, E.G.; Mouchiroud, L.; Cantó, C.; Fan, W.; Downes, M.; Héligon, C.; Barish, G.D.; Desvergne, B.; Evans, R.M.; et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 2011, 147, 827–839. [Google Scholar] [CrossRef]
- NCoR1 builds bigger muscles in mice. Bonekey Rep. 2012, 1, 15. [CrossRef]
- Opdenbosch, V.N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef]
- Fukuoka, M.; Fujita, H.; Numao, K.; Nakamura, Y.; Shimizu, H.; Sekiguchi, M.; Hohjoh, H. MiR-199-3p enhances muscle regeneration and ameliorates aged muscle and muscular dystrophy. Commun. Biol. 2021, 4, 427. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yu, M.; Kuma, A.; Klein, J.D.; Wang, Y.; Hassounah, F.; Cai, H.; Wang, X.H. Downregulation of let-7 by electrical acupuncture increases protein synthesis in mice. Front. Physiol. 2021, 12, 697139. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Tang, S.; Du, F.; Li, H.; Shen, X.; Li, D.; Wang, Y.; Zhang, Z.; Xia, L.; Zhu, Q.; et al. MiR-99a-5p regulates the proliferation and differentiation of skeletal muscle satellite cells by targeting MTMR3 in chicken. Genes 2020, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhu, J.; Zhang, R.; Liang, W.; Ma, W.; Zhang, Q.; Huang, Z.; Ding, F.; Sun, H. MiR-125b-5p targeting TRAF6 relieves skeletal muscle atrophy induced by fasting or denervation. Ann. Transl. Med. 2019, 7, 456. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, J. MicroRNA-let-7 Targets HMGA2 to Regulate the Proliferation, Migration, and Invasion of Colon Cancer Cell HCT116. Evid. Based Complement Alternat. Med. 2021, 2021, 2134942. [Google Scholar] [CrossRef]
- Wang, W.; Zhan, R.; Zhou, J.; Wang, J.; Chen, S. MiR-10 targets NgR to modulate the proliferation of microglial cells and the secretion of inflammatory cytokines. Exp. Mol. Pathol. 2018, 105, 357–363. [Google Scholar] [CrossRef]
- Peng, T.; Peng, J.J.; Miao, G.Y.; Tan, Z.Q.; Liu, B.; Zhou, E. MiR-125/CDK2 axis in cochlear progenitor cell proliferation. Mol. Med. Rep. 2021, 23, 102. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, R.; Lv, Y.; Dai, J.; Zhang, D.; Zhu, L.; Lin, Y. chi-miR-99b-3p Regulates the Proliferation of Goat Skeletal Muscle Satellite Cells In Vitro by Targeting Caspase-3 and NCOR1. Animals 2022, 12, 2368. https://doi.org/10.3390/ani12182368
Liao R, Lv Y, Dai J, Zhang D, Zhu L, Lin Y. chi-miR-99b-3p Regulates the Proliferation of Goat Skeletal Muscle Satellite Cells In Vitro by Targeting Caspase-3 and NCOR1. Animals. 2022; 12(18):2368. https://doi.org/10.3390/ani12182368
Chicago/Turabian StyleLiao, Rongrong, Yuhua Lv, Jianjun Dai, Defu Zhang, Lihui Zhu, and Yuexia Lin. 2022. "chi-miR-99b-3p Regulates the Proliferation of Goat Skeletal Muscle Satellite Cells In Vitro by Targeting Caspase-3 and NCOR1" Animals 12, no. 18: 2368. https://doi.org/10.3390/ani12182368
APA StyleLiao, R., Lv, Y., Dai, J., Zhang, D., Zhu, L., & Lin, Y. (2022). chi-miR-99b-3p Regulates the Proliferation of Goat Skeletal Muscle Satellite Cells In Vitro by Targeting Caspase-3 and NCOR1. Animals, 12(18), 2368. https://doi.org/10.3390/ani12182368




