Transcriptome Profile Analysis of Intestinal Upper Villus Epithelial Cells and Crypt Epithelial Cells of Suckling Piglets
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and CVA Cell Isolation
2.2. Measurement of Alkaline Phosphatase, Lactase, and Sucrase Activities
2.3. Total RNA Isolation
2.4. Processing of High-Throughput Sequencing Data
2.5. Differential Expression Analysis
2.6. Cluster Analysis and Functional Annotation of DEGs
2.7. qRT-PCR Validation of the Transcriptome Results
2.8. Statistical Analyses
3. Results
3.1. Enzyme Activities of the Fractionation Procedure
3.2. Mapping of RNA-Seq Reads to Pig Genome
3.3. Differential Gene Expression Profiles of F1 and F3
3.4. GO and KEGG Pathway Analyses of DEGs
3.5. Experimental Validation of DEGs Expression Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, G. Intestinal mucosal amino acid catabolism. J. Nutr. 1998, 128, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Xiong, X.; Wang, X.; Yang, H.; Li, J.; Huang, P.; Long, Y. Current advances in renewal mechanisms of intestinal epithelial cells along the crypt-villus axis. Sci. Sin. Vitae 2017, 47, 190–200. [Google Scholar]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Xiong, X.; Yang, H.; Hu, X.; Wang, X.; Li, B.; Long, L.; Li, T.; Wang, J.; Hou, Y.; Wu, G.; et al. Differential proteome analysis along jejunal crypt-villus axis in piglets. Front. Biosci. 2016, 21, 343–363. [Google Scholar]
- Van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef]
- Zhu, L.H.; Xu, J.X.; Zhu, S.W.; Cai, X.; Yang, S.F.; Chen, X.L.; Guo, Q. Gene expression profiling analysis reveals weaning-induced cell cycle arrest and apoptosis in the small intestine of pigs. J. Anim. Sci. 2014, 92, 996–1006. [Google Scholar] [CrossRef]
- Xiong, X.; Yang, H.; Tan, B.; Yang, C.; Wu, M.; Liu, G.; Kim, S.W.; Li, T.; Li, L.; Wang, J.; et al. Differential expression of proteins involved in energy production along the crypt-villus axis in early-weaning pig small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G229–G237. [Google Scholar] [CrossRef]
- Zou, L.; Xiong, X.; Yang, H.; Wang, K.; Zhou, J.; Lv, D.; Yin, Y. Identification of microRNA transcriptome reveals that miR-100 is involved in the renewal of porcine intestinal epithelial cells. Sci. China Life Sci. 2019, 62, 816–828. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Moeser, A.J.; Ryan, K.A.; Nighot, P.K.; Blikslager, A.T. Gastrointestinal dysfunction induced by early weaning is attenuated by delayed weaning and mast cell blockade in pigs. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G413–G421. [Google Scholar] [CrossRef] [PubMed]
- Lackeyram, D.; Yang, C.; Archbold, T.; Swanson, K.C.; Fan, M.Z. Early weaning reduces small intestinal alkaline phosphatase expression in pigs. J. Nutr. 2010, 140, 461–468. [Google Scholar] [CrossRef]
- Yang, H.; Xiong, X.; Wang, X.; Tan, B.; Li, T.; Yin, Y. Effects of Weaning on Intestinal Upper Villus Epithelial Cells of Piglets. PLoS ONE 2016, 11, e0150216. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.H.; Xiao, K.; Luan, Z.S.; Song, J. Early weaning increases intestinal permeability, alters expression of cytokine and tight junction proteins, and activates mitogen-activated protein kinases in pigs. J. Anim. Sci. 2013, 91, 1094–1101. [Google Scholar] [CrossRef]
- Diakogiannaki, E.; Pais, R.; Tolhurst, G.; Parker, H.E.; Horscroft, J.; Rauscher, B.; Zietek, T.; Daniel, H.; Gribble, F.M.; Reimann, F. Oligopeptides stimulate glucagon-like peptide-1 secretion in mice through proton-coupled uptake and the calcium-sensing receptor. Diabetologia 2013, 56, 2688–2696. [Google Scholar] [CrossRef]
- Fawkner-Corbett, D.; Antanaviciute, A.; Parikh, K.; Jagielowicz, M.; Geros, A.S.; Gupta, T.; Ashley, N.; Khamis, D.; Fowler, D.; Morrissey, E.; et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 2021, 184, 810–826. [Google Scholar] [CrossRef]
- Pu, Y.; Li, S.; Xiong, H.; Zhang, X.; Wang, Y.; Du, H. Iron Promotes Intestinal Development in Neonatal Piglets. Nutrients 2018, 10, 726. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J.; Wreschner, D.H.; Tran, M.; Eyre, H.J.; Sutherland, G.R.; McGuckin, M.A. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J. Biol. Chem. 2001, 276, 18327–18336. [Google Scholar] [CrossRef] [PubMed]
- Sancho, R.; Nateri, A.S.; de Vinuesa, A.G.; Aguilera, C.; Nye, E.; Spencer-Dene, B.; Behrens, A. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 2009, 28, 1843–1854. [Google Scholar] [CrossRef]
- Biteau, B.; Jasper, H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 2011, 138, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Edgar, B.A.; Boutros, M. ATF3 acts as a rheostat to control JNK signalling during intestinal regeneration. Nat. Commun. 2017, 8, 14289. [Google Scholar] [CrossRef]
- Yu, T.; Chen, X.; Zhang, W.; Li, J.; Xu, R.; Wang, T.C.; Ai, W.; Liu, C. Kruppel-like factor 4 regulates intestinal epithelial cell morphology and polarity. PLoS ONE 2012, 7, e32492. [Google Scholar] [CrossRef] [PubMed]
- Dunker, N.; Schmitt, K.; Schuster, N.; Krieglstein, K. The role of transforming growth factor beta-2, beta-3 in mediating apoptosis in the murine intestinal mucosa. Gastroenterology 2002, 122, 1364–1375. [Google Scholar] [CrossRef]
- McKaig, B.C.; Makh, S.S.; Hawkey, C.J.; Podolsky, D.K.; Mahida, Y.R. Normal human colonic subepithelial myofibroblasts enhance epithelial migration (restitution) via TGF-beta3. Am. J. Physiol. 1999, 276, G1087–G1093. [Google Scholar] [PubMed]
- Chang, J.; Cizmecioglu, O.; Hoffmann, I.; Rhee, K. PLK2 phosphorylation is critical for CPAP function in procentriole formation during the centrosome cycle. EMBO J. 2010, 29, 2395–2406. [Google Scholar] [CrossRef]
- Katoh, M. WNT signaling in stem cell biology and regenerative medicine. Curr. Drug Targets 2008, 9, 565–570. [Google Scholar] [CrossRef]
- Wend, P.; Wend, K.; Krum, S.A.; Miranda-Carboni, G.A. The role of WNT10B in physiology and disease. Acta Physiol. 2012, 204, 34–51. [Google Scholar] [CrossRef]
- Batlle, E.; Henderson, J.T.; Beghtel, H.; van den Born, M.M.; Sancho, E.; Huls, G.; Meeldijk, J.; Robertson, J.; van de Wetering, M.; Pawson, T.; et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 2002, 111, 251–263. [Google Scholar] [CrossRef]
- Hishida, R.; Ishihara, T.; Kondo, K.; Katsura, I. hch-1, a gene required for normal hatching and normal migration of a neuroblast in C. elegans, encodes a protein related to TOLLOID and BMP-1. EMBO J. 1996, 15, 4111–4122. [Google Scholar] [CrossRef]
- He, X.C.; Zhang, J.; Tong, W.G.; Tawfik, O.; Ross, J.; Scoville, D.H.; Tian, Q.; Zeng, X.; He, X.; Wiedemann, L.M.; et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004, 36, 1117–1121. [Google Scholar] [CrossRef]
- Auclair, B.A.; Benoit, Y.D.; Rivard, N.; Mishina, Y.; Perreault, N. Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology 2007, 133, 887–896. [Google Scholar] [CrossRef]
- Snippert, H.J.; van der Flier, L.G.; Sato, T.; van Es, J.H.; van den Born, M.; Kroon-Veenboer, C.; Barker, N.; Klein, A.M.; van Rheenen, J.; Simons, B.D.; et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010, 143, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Ordonez-Moran, P.; Dafflon, C.; Imajo, M.; Nishida, E.; Huelsken, J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell 2015, 28, 815–829. [Google Scholar] [CrossRef]
- Imajo, M.; Ebisuya, M.; Nishida, E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat. Cell Biol. 2015, 17, 7–19. [Google Scholar] [CrossRef]
- Ding, Q.; Wang, Q.; Evers, B.M. Alterations of MAPK activities associated with intestinal cell differentiation. Biochem. Biophys. Res. Commun. 2001, 284, 282–288. [Google Scholar] [CrossRef]
- Zhang, F.; Pirooznia, M.; Xu, H. Mitochondria regulate intestinal stem cell proliferation and epithelial homeostasis through FOXO. Mol. Biol. Cell 2020, 31, 1538–1549. [Google Scholar] [CrossRef]
- Ling, M.; Quan, L.; Lai, X.; Lang, L.; Li, F.; Yang, X.; Fu, Y.; Feng, S.; Yi, X.; Zhu, C.; et al. VEGFB Promotes Myoblasts Proliferation and Differentiation through VEGFR1-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2021, 22, 13352. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Fritsch, S.D.; Weichhart, T. Metabolic and immunologic control of intestinal cell function by mTOR. Int. Immunol. 2020, 32, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, J.; Kotani, T.; Konno, T.; Saito, Y.; Murata, Y.; Noda, T.; Matozaki, T. Regulation of Small Intestinal Epithelial Homeostasis by Tsc2-mTORC1 Signaling. Kobe J. Med. Sci. 2019, 64, E200–E209. [Google Scholar]
- Sun, X.; Du, M.; Navarre, D.A.; Zhu, M.J. Purple Potato Extract Promotes Intestinal Epithelial Differentiation and Barrier Function by Activating AMP-Activated Protein Kinase. Mol. Nutr. Food Res. 2018, 62, 1700536. [Google Scholar] [CrossRef] [PubMed]
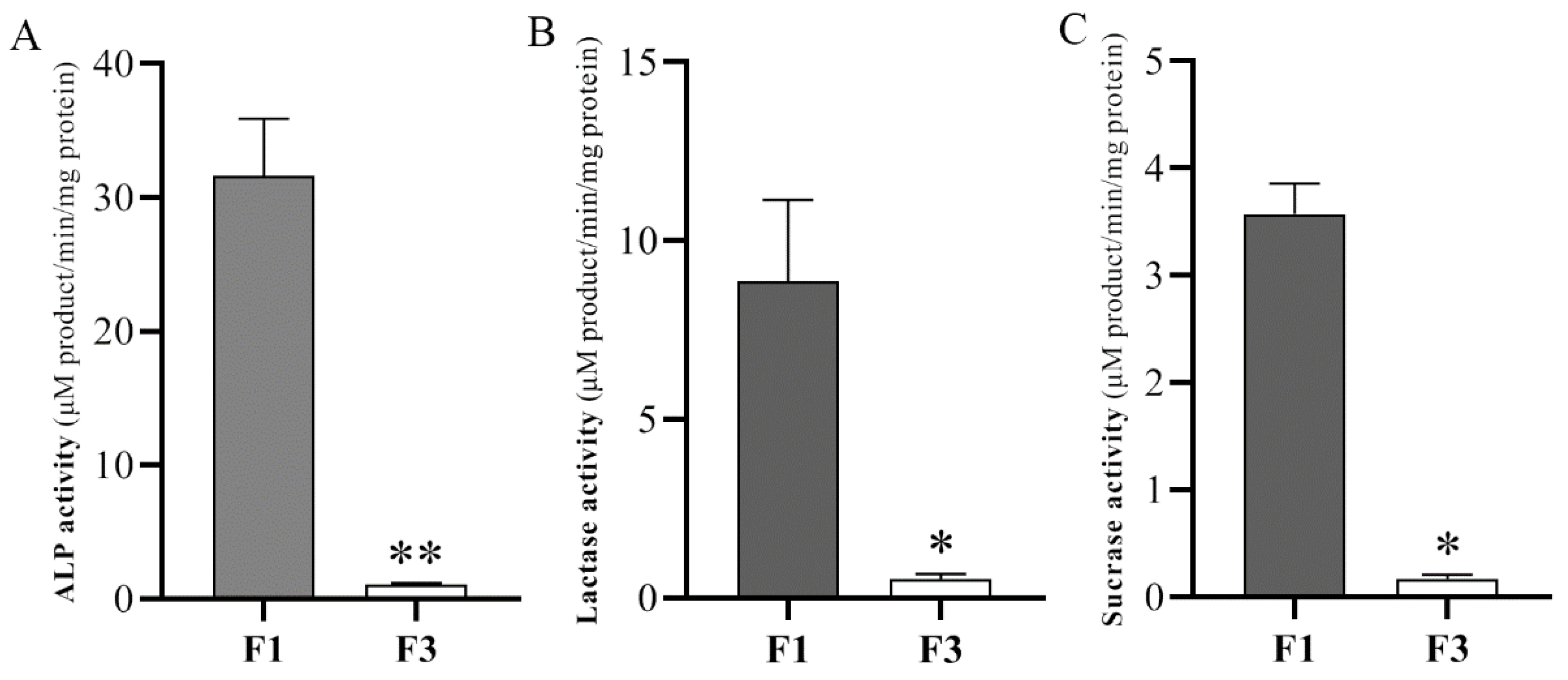
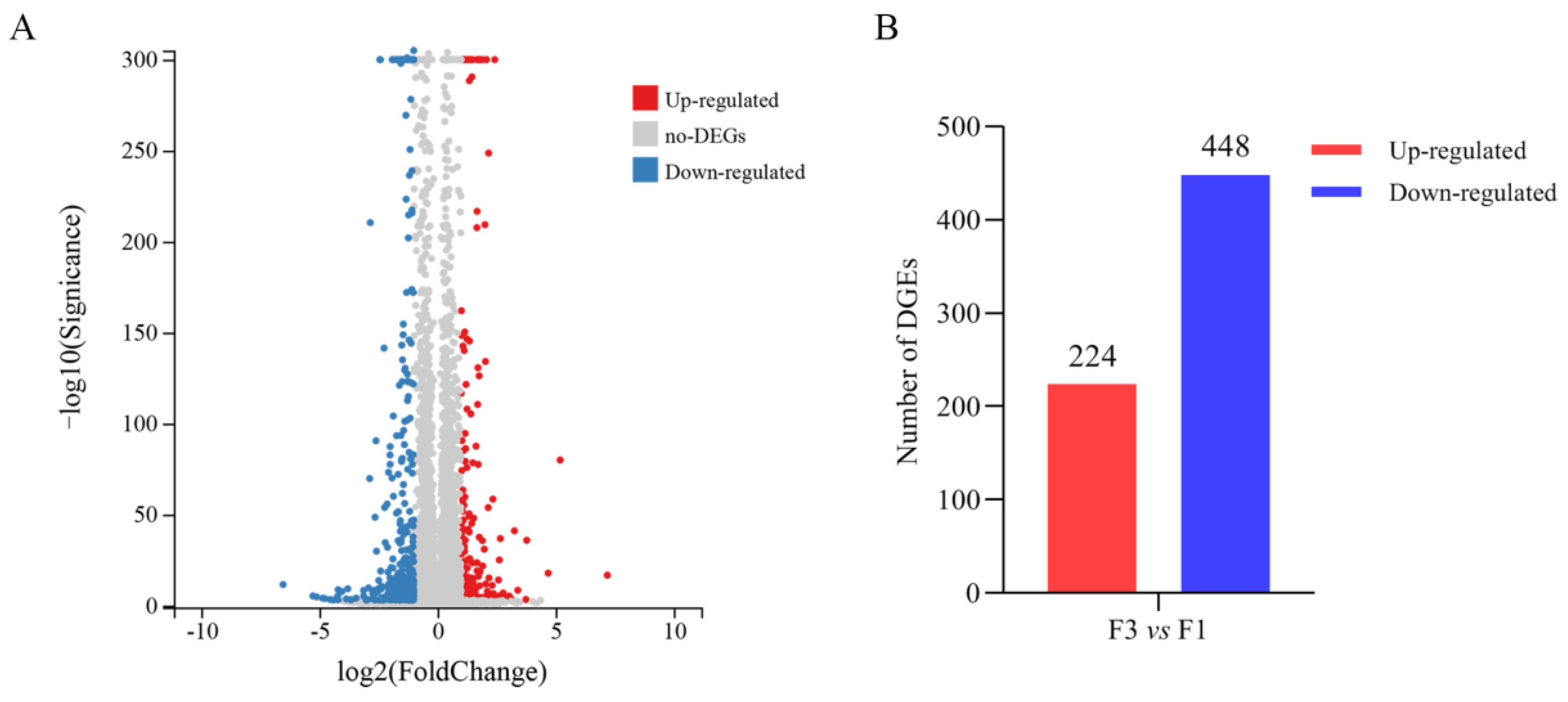


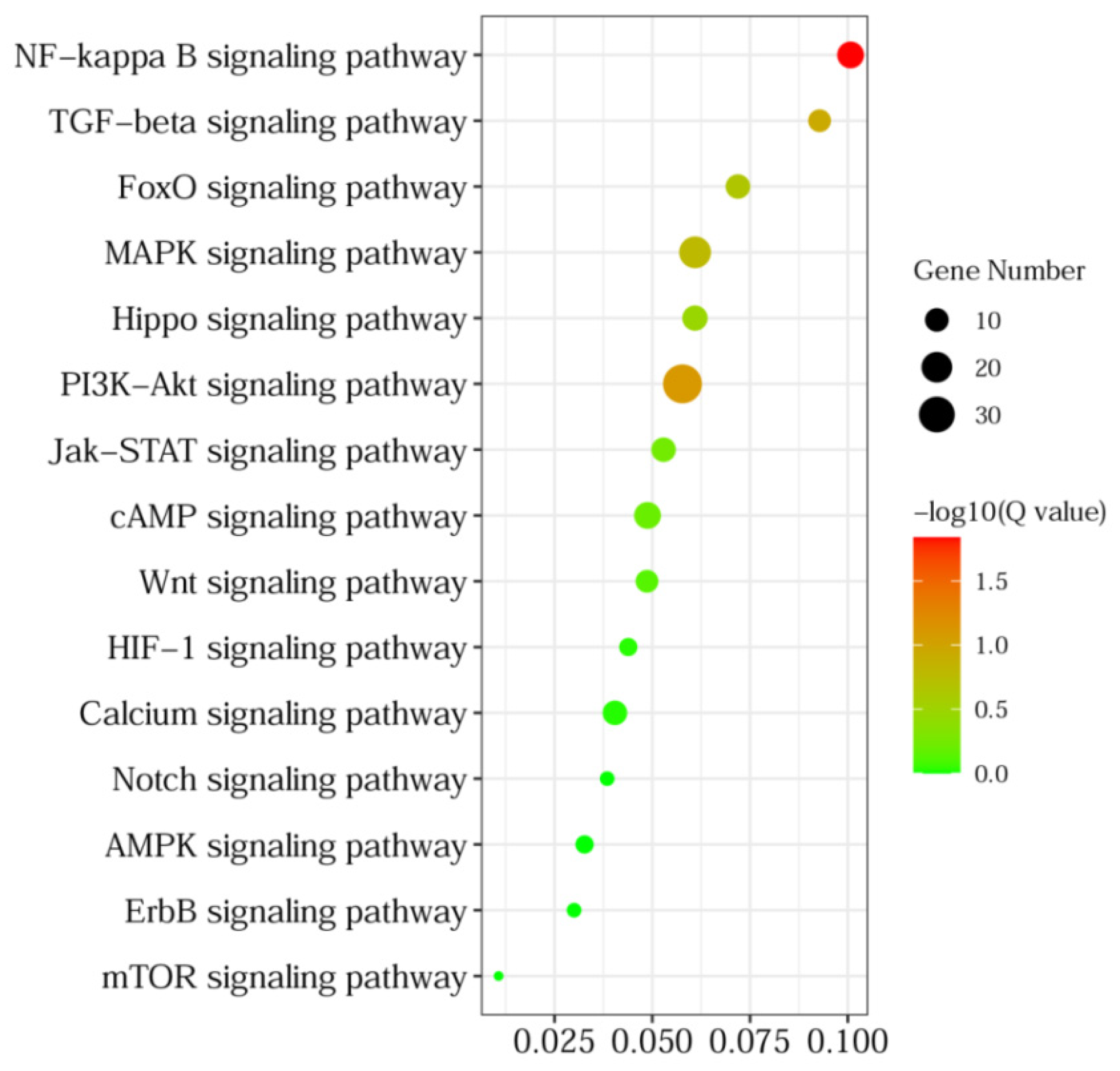
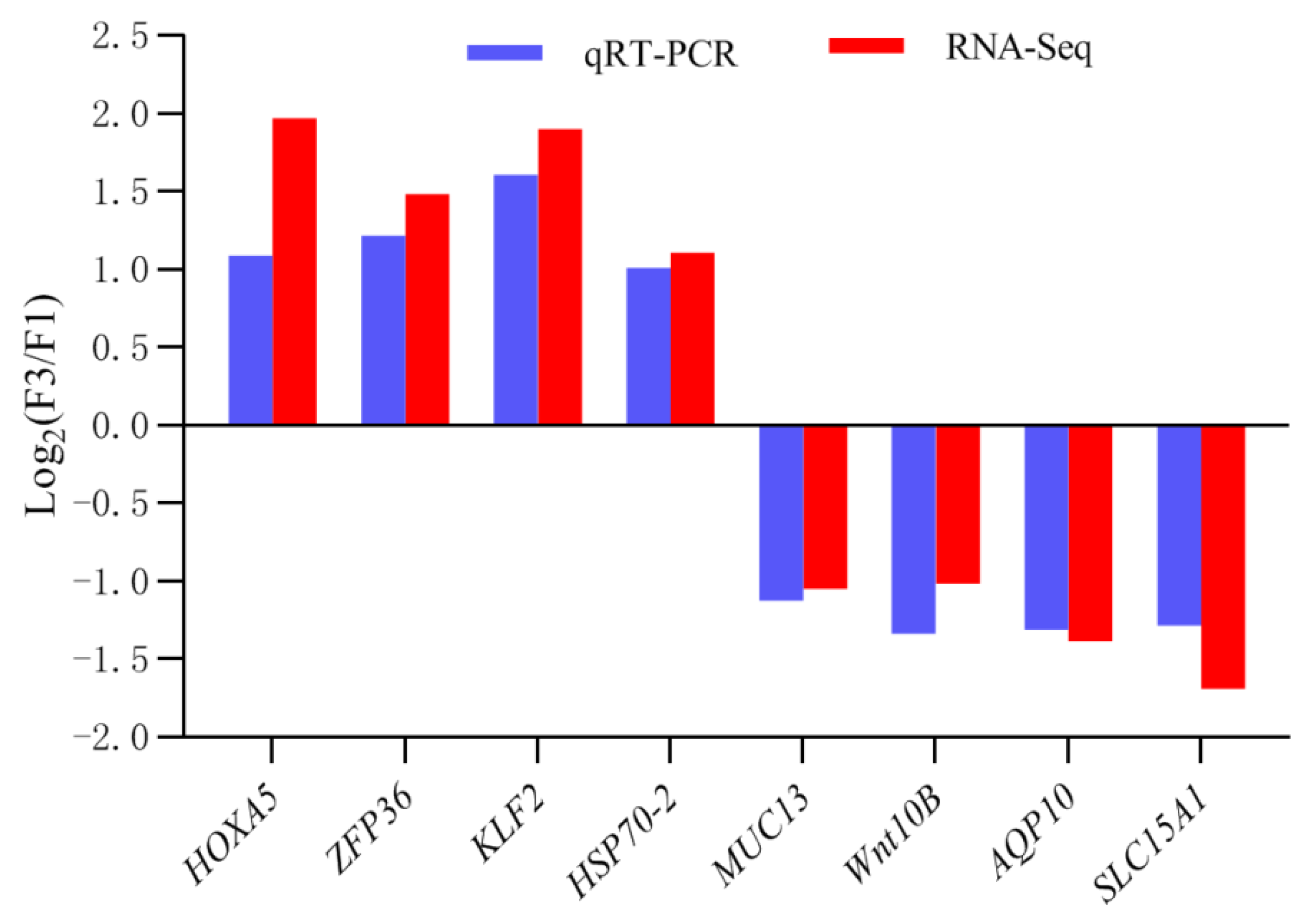

| Sample Name | F1 a | F3 b |
|---|---|---|
| Raw Reads | 23,957,221 | 23,957,192 |
| Q20 c | 93.90 | 96.00 |
| Clean Reads | 23,881,637 | 23,894,452 |
| Total Mapped Reads (%) | 88.35 | 90.21 |
| Unique Match (%) | 57.16 | 63.18 |
| Multi-position Match (%) | 31.19 | 27.03 |
| Total Unmapped Reads (%) | 11.65 | 9.80 |
| Gene ID | Symbol | Description (Gene Name) | Log2 Ratio(F3/F1) ab | FDR c |
|---|---|---|---|---|
| 397113 | SLC5A1 | Sodium/Glucose Cotransporter 1 (SGLT1) | −1.149 | 0 |
| 397624 | SLC15A1 | Peptide-transporters 1 (PEPT1) | −1.691 | 0 |
| 100144588 | SLC3A1 | Neutral And Basic Amino Acid Transporter (NBAT) | −1.385 | 5.17 × 10−3 |
| 100520648 | SLC16A8 | Monocarboxylate Transporter 3 (MCT3) | −1.765 | 8.19 × 10−4 |
| 100737517 | SLC40A1 | Iron-Regulated Transporter 1 (IRT1) | −1.144 | 0 |
| 396845 | TF | Transferrin | −1.222 | 1.465 × 10−20 |
| 100125829 | MUC13 | Mucin 13 | −1.050 | 0 |
| 100126276 | WNT10B | Wingless-Type MMTV Integration Site Family, Member 10B | −1.015 | 3.028 × 10−6 |
| 100156461 | BMP1 | Bone Morphogenetic Protein 1 | −1.915 | 0 |
| 100151994 | LGR5 | Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 | 1.031 | 8.174 × 10−30 |
| 397423 | HOXA5 | Homeobox A5 | 1.969 | 2.04 × 10−5 |
| 396913 | JUN | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit (c-Jun) | 1.362 | 0 |
| 100144486 | FOS | Fos Proto-Oncogene, AP-1 Transcription Factor Subunit (c-Fos) | 1.769 | 0 |
| 100520726 | EGR1 | Early Growth Response 1 | 2.074 | 0 |
| 100738612 | ATF3 | Activating Transcription Factor 3 | 1.875 | 0 |
| 595111 | KLF4 | Krüppel-Like Factor 4 | 1.161 | 0 |
| 397251 | PLK2 | Polo-Like Kinase 2 | 2.058 | 0 |
| 397400 | TGFB3 | Transforming Growth Factor Beta 3 | 1.607 | 7.343 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, L.; Shao, Y.; Xu, Y.; Wu, Y.; Zhou, J.; Xiong, X.; Yin, Y. Transcriptome Profile Analysis of Intestinal Upper Villus Epithelial Cells and Crypt Epithelial Cells of Suckling Piglets. Animals 2022, 12, 2324. https://doi.org/10.3390/ani12182324
Zou L, Shao Y, Xu Y, Wu Y, Zhou J, Xiong X, Yin Y. Transcriptome Profile Analysis of Intestinal Upper Villus Epithelial Cells and Crypt Epithelial Cells of Suckling Piglets. Animals. 2022; 12(18):2324. https://doi.org/10.3390/ani12182324
Chicago/Turabian StyleZou, Lijun, Yirui Shao, Yinfeng Xu, Yuliang Wu, Jian Zhou, Xia Xiong, and Yulong Yin. 2022. "Transcriptome Profile Analysis of Intestinal Upper Villus Epithelial Cells and Crypt Epithelial Cells of Suckling Piglets" Animals 12, no. 18: 2324. https://doi.org/10.3390/ani12182324
APA StyleZou, L., Shao, Y., Xu, Y., Wu, Y., Zhou, J., Xiong, X., & Yin, Y. (2022). Transcriptome Profile Analysis of Intestinal Upper Villus Epithelial Cells and Crypt Epithelial Cells of Suckling Piglets. Animals, 12(18), 2324. https://doi.org/10.3390/ani12182324





