Three-Dimensional Investigations of Virus-Associated Structures in the Nuclei with White Spot Syndrome Virus (WSSV) Infection in Red Swamp Crayfish (Procambarus clarkii)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Virus Preparation and Negative Stain
2.3. Virus Inoculation and Perfusion Fixation
2.4. Ultrastructural Observation and Electron Tomography
2.5. Immuno-Gold Labeling
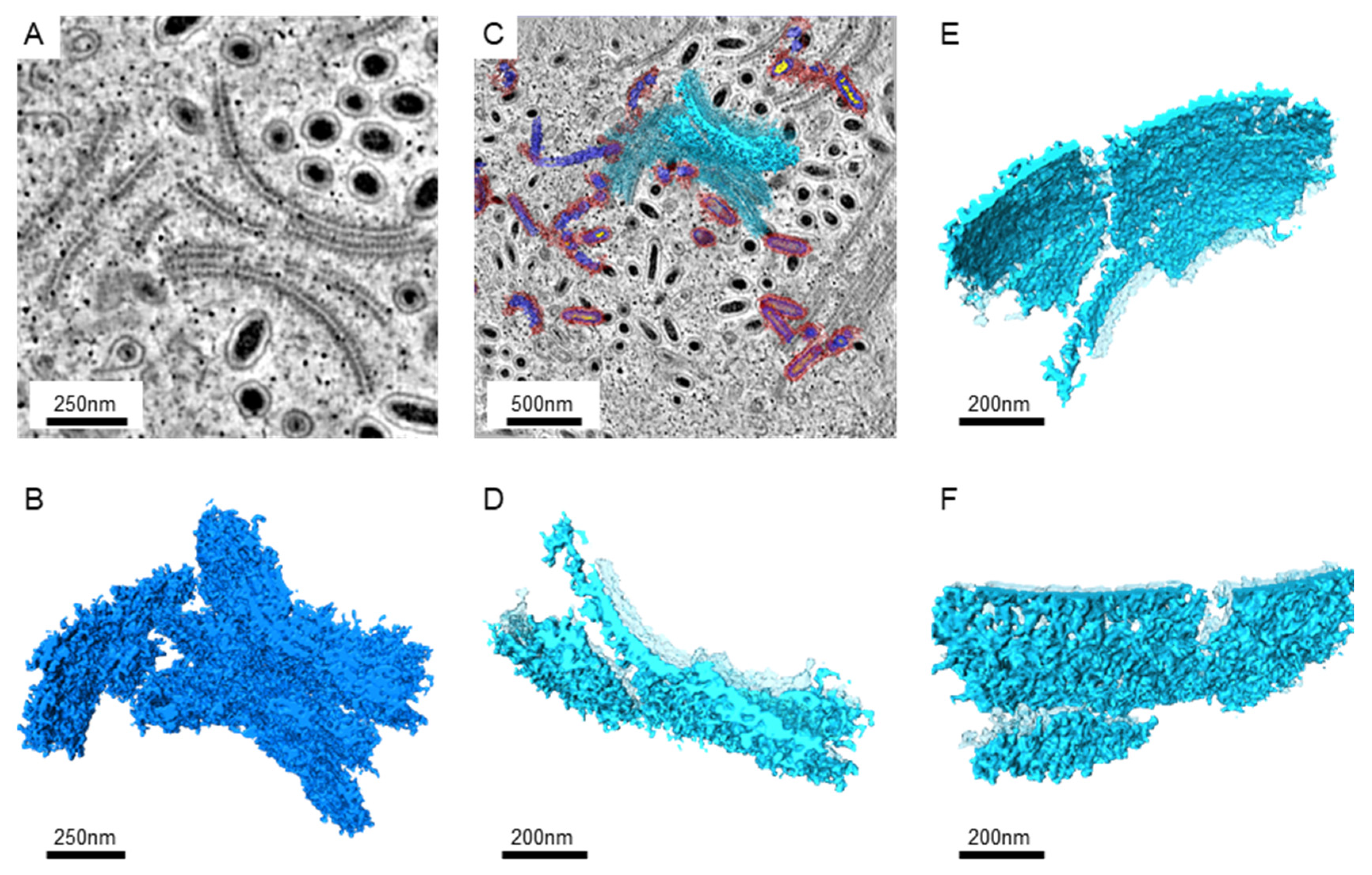
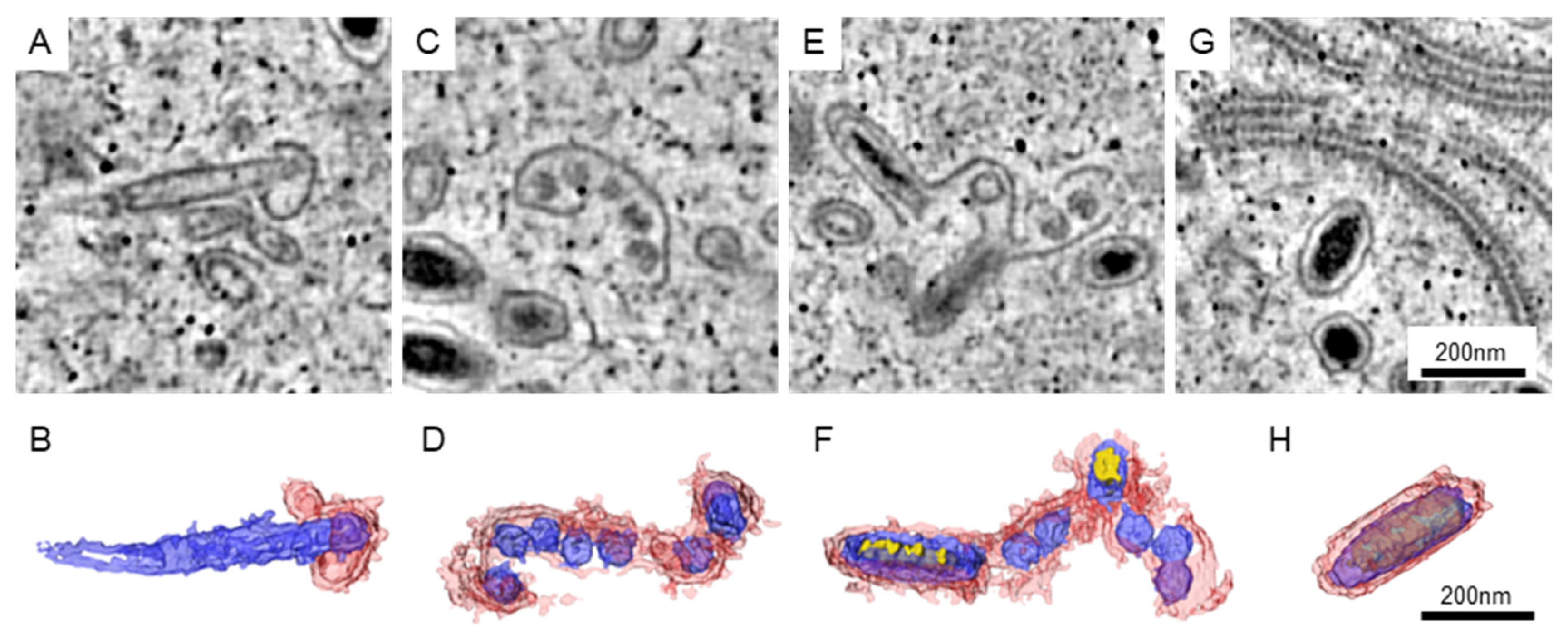
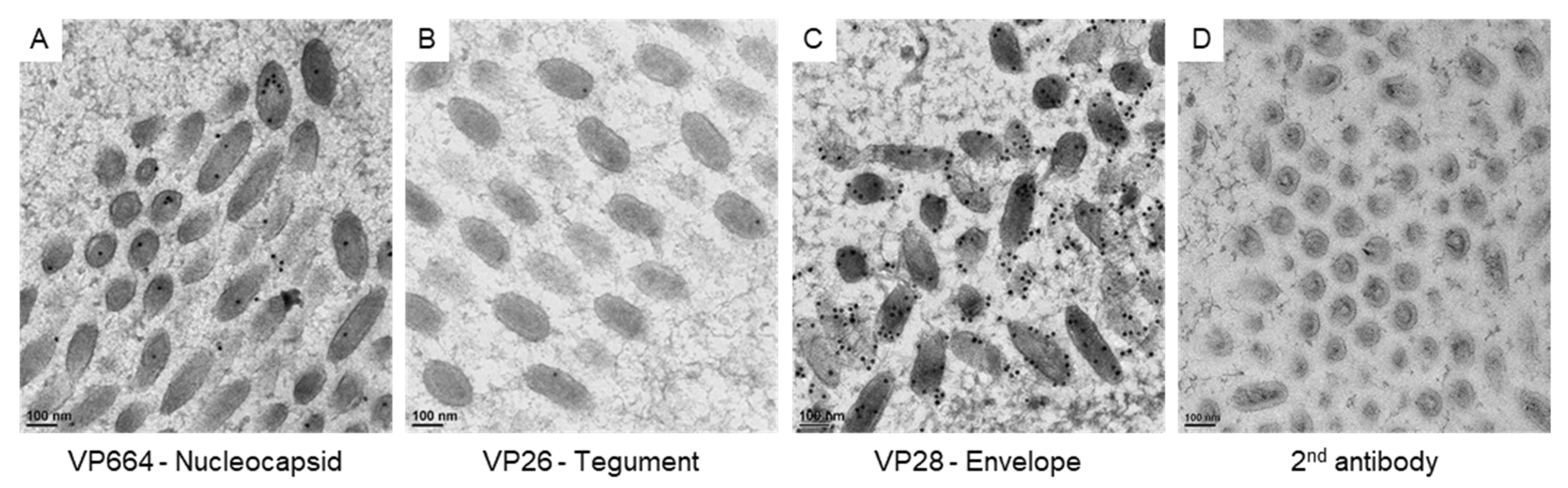
3. Results
3.1. The Ultrastructure of the WSSV
3.2. The Assembling Particles in WSSV-Infected Nucleus
3.3. The Protein Expression on the Virus-Associated Structures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chou Hy, H.C.Y.; Wang, C.H.; Chiang, H.C.; Lo, C.F. Pathogenicity of a baculovirus infection causing white spot syndrome in cultured penaeid shrimp in Taiwan. Dis. Aquat. Org. 1995, 23, 165–173. [Google Scholar] [CrossRef]
- Stentiford, G.D.; Bonami, J.R.; Alday-Sanz, V. A critical review of susceptibility of crustaceans to Taura syndrome, Yellowhead disease and White Spot Disease and implications of inclusion of these diseases in European legislation. Aquaculture 2009, 291, 1–17. [Google Scholar] [CrossRef]
- Tran, N.T.; Liang, H.; Zhang, M.; Bakky, M.A.H.; Zhang, Y.; Li, S. Role of cellular receptors in the innate immune system of crustaceans in response to white spot syndrome virus. Viruses 2022, 14, 743. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.H.; Yang, F.; Zhang, X.; Xu, X.; Kou, G.H.; Lo, C.F. Whispovirus. In Lesser Known Large dsDNA Viruses, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 197–227. [Google Scholar]
- Verbruggen, B.; Bickley, L.K.; van Aerle, R.; Bateman, K.S.; Stentiford, G.D.; Santos, E.M.; Tyler, C.R. Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments. Viruses 2016, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Chen, L.L. WSSV envelope protein VP51B links structural protein complexes and may mediate virus infection. J. Fish Dis. 2017, 40, 571–581. [Google Scholar] [CrossRef]
- Escobedo-Bonilla, C.M.; Alday-Sanz, V.; Wille, M.; Sorgeloos, P.; Pensaert, M.B.; Nauwynck, H.J. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 2008, 31, 1–18. [Google Scholar] [CrossRef]
- Manjusha, M.; Varghese, R.; Philip, R.; Mohandas, A.; Bright Singh, I.S. Pathological changes in Fenneropenaeus indicus experimentally infected with white spot virus and virus morphogenesis. J. Invertebr. Pathol. 2009, 102, 225–232. [Google Scholar] [CrossRef]
- Wang, C.H.; Yang, H.N.; Tang, C.Y.; Lu, C.H.; Kou, G.H.; Lo, C.F. Ultrastructure of white spot syndrome virus development in primary lymphoid organ cell cultures. Dis. Aquat. Org. 2000, 41, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Durand, S.; Lightner, D.V.; Redman, R.M.; Bonami, J.R. Ultrastructure and morphogenesis of White Spot Syndrome Baculovirus (WSSV). Dis. Aquat. Org. 1997, 29, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.G.; Hassan, M.D.; Shariff, M.; Zamri, S.M.; Chen, X. Histopathology and cytopathology of white spot syndrome virus (WSSV) in cultured Penaeus monodon from peninsular Malaysia with emphasis on pathogenesis and the mechanism of white spot formation. Dis. Aquat. Org. 1999, 39, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Tsai, J.M.; Wang, H.C.; Leu, J.H.; Wang, A.H.; Zhuang, Y.; Walker, P.J.; Kou, G.H.; Lo, C.F. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 2006, 80, 3021–3029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Brey, I.; Bartenschlager, R. Viral infection at high magnification: 3D electron microscopy methods to analyze the architecture of infected cells. Viruses 2015, 7, 6316–6345. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hong, Y.; Huo, D.; Cai, P. Ultrastructure analysis of white spot syndrome virus (WSSV). Arch. Virol. 2020, 165, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hong, Y.; Qiu, H.; Yang, F.; Li, F. VP19 is important for the envelope coating of white spot syndrome virus. Virus Res. 2019, 270, 197666. [Google Scholar] [CrossRef]
- Romero-Brey, I. 3D electron microscopy (EM) and correlative light electron microscopy (CLEM) methods to study virus-host interactions. In Influenza Virus: Methods and Protocols, 1st ed.; Humana: New York, NY, USA, 2018; pp. 213–236. [Google Scholar]
- Freire, C.A.; McNamara, J.C. Fine structure of the gills of the fresh-water shrimp Macrobrachium olfersii (Decapoda): Effect of acclimation to high salinity medium and evidence for involvement of the lamellar septum in ion uptake. J. Crustac. Biol. 1995, 15, 103–116. [Google Scholar] [CrossRef]
- Jiang, Y.F.; Lin, H.L.; Fu, C.Y. 3D mitochondrial ultrastructure of Drosophila indirect flight muscle Revealed by serial-section electron tomography. J. Vis. Exp. 2017, 130, e56567. [Google Scholar] [CrossRef] [Green Version]
- Mastronarde, D.N.; Held, S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 2017, 197, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Leu, J.H.; Tsai, J.M.; Wang, H.C.; Wang, A.H.; Wang, C.H.; Kou, G.H.; Lo, C.F. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J. Virol. 2005, 79, 140–149. [Google Scholar] [CrossRef] [Green Version]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of virus assembly. Annu. Rev. Phys. Chem. 2015, 66, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Durand, S.; Lightner, D.V.; Nunan, L.M.; Redman, R.M.; Mari, J.; Bonami, J.R. Application of gene probes as diagnostic tools for White Spot Baculovirus (WSBV) of penaeid shrimp. Dis. Aquat. Org. 1996, 27, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Van Hulten, M.C.; Witteveldt, J.; Peters, S.; Kloosterboer, N.; Tarchini, R.; Fiers, M.; Sandbrink, H.; Lankhorst, R.K.; Vlak, J.M. The white spot syndrome virus DNA genome sequence. Virology 2001, 286, 7–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Lin, Q.; Chen, J.; Wu, J.L.; Lim, T.K.; Loh, S.S.; Tang, X.; Hew, C.L. Shotgun identification of the structural proteome of shrimp white spot syndrome virus and iTRAQ differentiation of envelope and nucleocapsid subproteomes. Mol. Cell. Proteom. 2007, 6, 1609–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskaran Sathyabhama, A.; Puthumana, J.; Kombiyil, S.; Philip, R.; Bright Singh, I.S. ‘PmLyO-Sf9—WSSV complex’ could be a platform for elucidating the mechanism of viral entry, cellular apoptosis and replication impediments. Virology 2021, 553, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, K.; Kumar, P.P.; Praveenraj, J.; Baruah, A.; Sivaramakrishnan, T.; Kumar, T.S.; Kumar, S.P.; Sankar, R.K.; Roy, S.D. Investigation and confirmation of white spot syndrome virus (WSSV) infection in wild caught penaeid shrimps of Andaman and Nicobar Islands, India. Virusdisease 2017, 28, 368–372. [Google Scholar] [CrossRef]
- Rodriguez, J.; Bayot, B.; Amano, Y.; Panchana, F.; de Blas, I.; Alday, V.; Calderon, J. White spot syndrome virus infection in cultured Penaeus vannamei (Boone) in Ecuador with emphasis on histopathology and ultrastructure. J. Fish Dis. 2003, 26, 439–450. [Google Scholar] [CrossRef]
- Zlotnick, A.; Johnson, J.M.; Wingfield, P.W.; Stahl, S.J.; Endres, D. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry 1999, 38, 14644–14652. [Google Scholar] [CrossRef]
- Goicochea, N.L.; De, M.; Rotello, V.M.; Mukhopadhyay, S.; Dragnea, B. Core-like particles of an enveloped animal virus can self-assemble efficiently on artificial templates. Nano Lett. 2007, 7, 2281–2290. [Google Scholar] [CrossRef]
- Kler, S.; Asor, R.; Li, C.; Ginsburg, A.; Harries, D.; Oppenheim, A.; Zlotnick, A.; Raviv, U. RNA encapsidation by SV40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 2012, 134, 8823–8830. [Google Scholar] [CrossRef] [Green Version]
- Welsch, S.; Muller, B.; Krausslich, H.G. More than one door—Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007, 581, 2089–2097. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Boura, E.; Carlson, L.A.; Rozycki, B. Membrane budding. Cell 2010, 143, 875–887. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.P.; Koyuncu, O.O.; Enquist, L.W. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011, 9, 427–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gladnikoff, M.; Shimoni, E.; Gov, N.S.; Rousso, I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys. J. 2009, 97, 2419–2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]


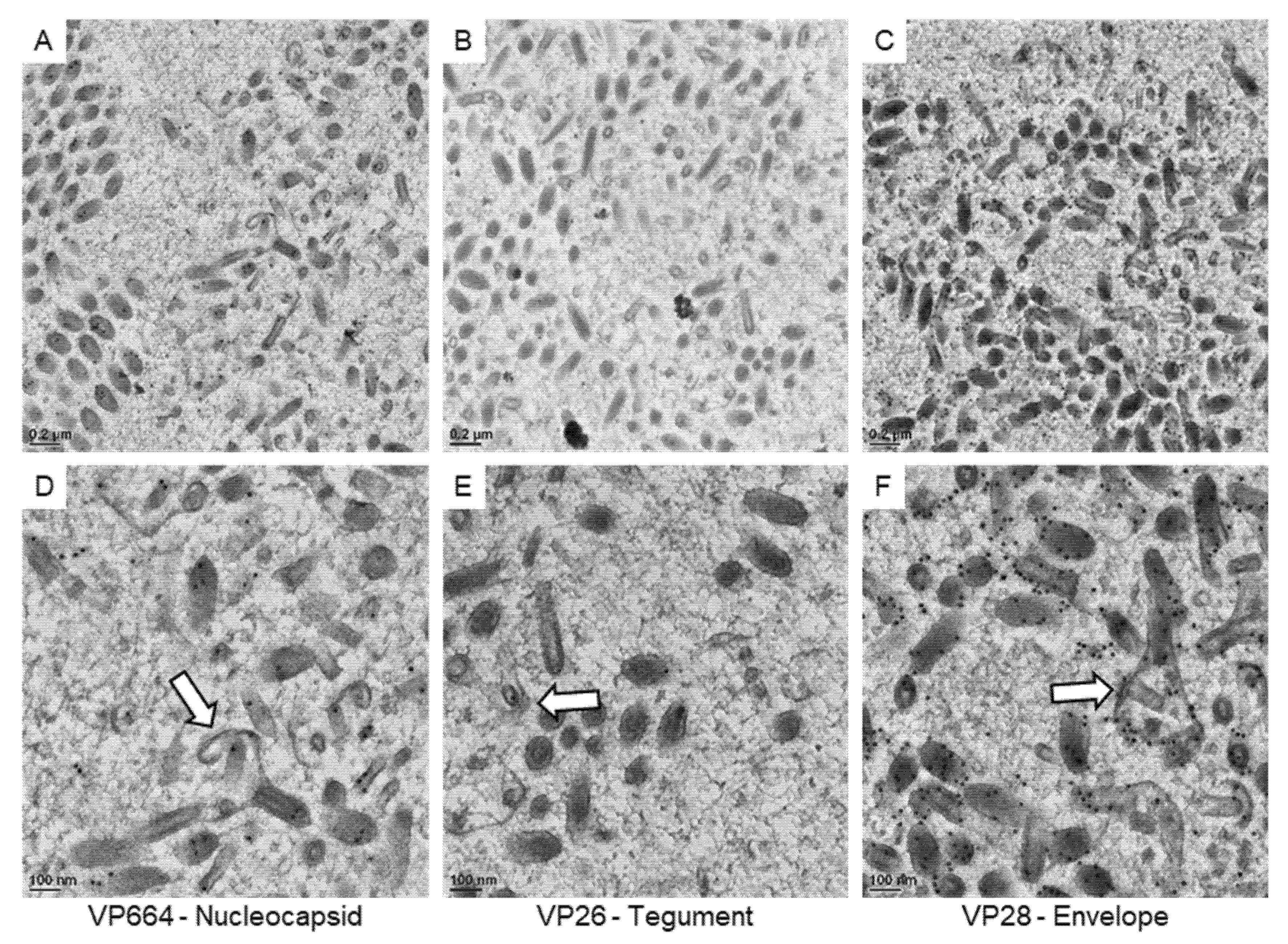
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budi, Y.P.; Lin, L.-C.; Chung, C.-H.; Chen, L.-L.; Jiang, Y.-F. Three-Dimensional Investigations of Virus-Associated Structures in the Nuclei with White Spot Syndrome Virus (WSSV) Infection in Red Swamp Crayfish (Procambarus clarkii). Animals 2022, 12, 1730. https://doi.org/10.3390/ani12131730
Budi YP, Lin L-C, Chung C-H, Chen L-L, Jiang Y-F. Three-Dimensional Investigations of Virus-Associated Structures in the Nuclei with White Spot Syndrome Virus (WSSV) Infection in Red Swamp Crayfish (Procambarus clarkii). Animals. 2022; 12(13):1730. https://doi.org/10.3390/ani12131730
Chicago/Turabian StyleBudi, Yovita Permata, Li-Chi Lin, Chang-Hsien Chung, Li-Li Chen, and Yi-Fan Jiang. 2022. "Three-Dimensional Investigations of Virus-Associated Structures in the Nuclei with White Spot Syndrome Virus (WSSV) Infection in Red Swamp Crayfish (Procambarus clarkii)" Animals 12, no. 13: 1730. https://doi.org/10.3390/ani12131730
APA StyleBudi, Y. P., Lin, L.-C., Chung, C.-H., Chen, L.-L., & Jiang, Y.-F. (2022). Three-Dimensional Investigations of Virus-Associated Structures in the Nuclei with White Spot Syndrome Virus (WSSV) Infection in Red Swamp Crayfish (Procambarus clarkii). Animals, 12(13), 1730. https://doi.org/10.3390/ani12131730






