Polymorphisms of the Growth Hormone Releasing Hormone Receptor Gene Affect Body Conformation Traits in Chinese Dabieshan Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
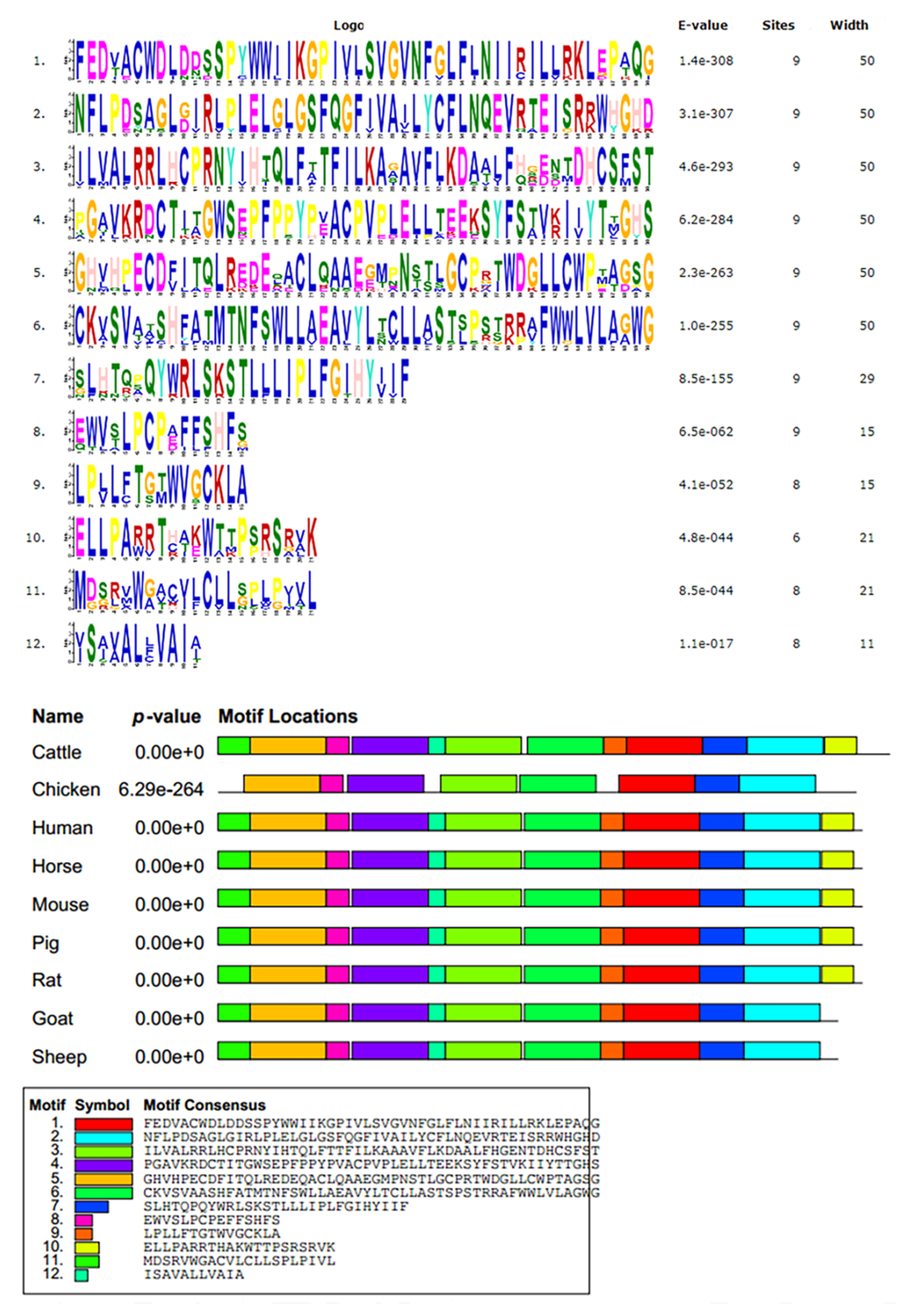
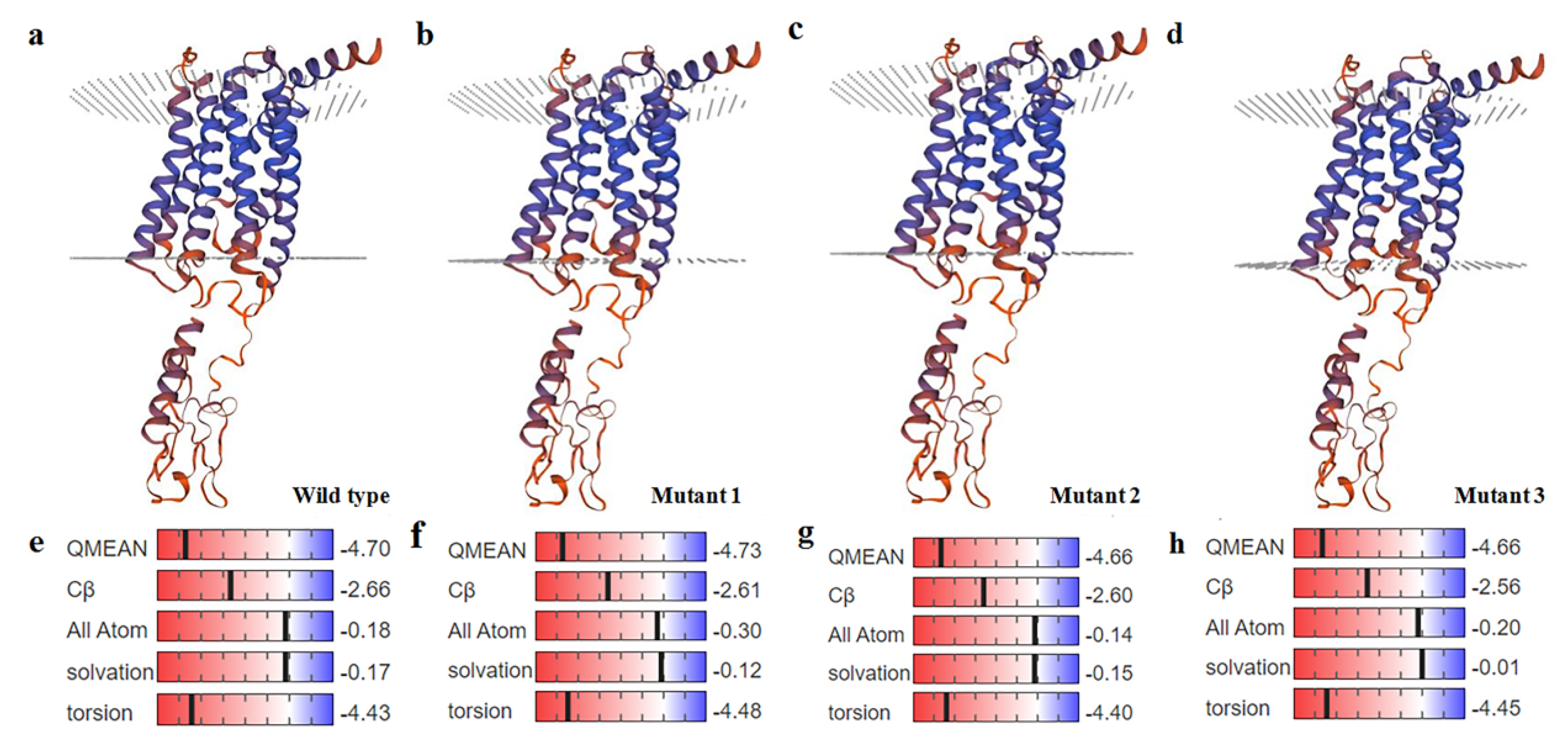
2.2. Sequence Alignment, Phylogenetic Analysis, Motif Analysis, and Predicted Structures
2.3. Phenotypic Data and DNA Sample Collection
2.4. Primers, Polymerase Chain Reaction, and Gel Electrophoresis
2.5. Statistical Analysis
3. Results
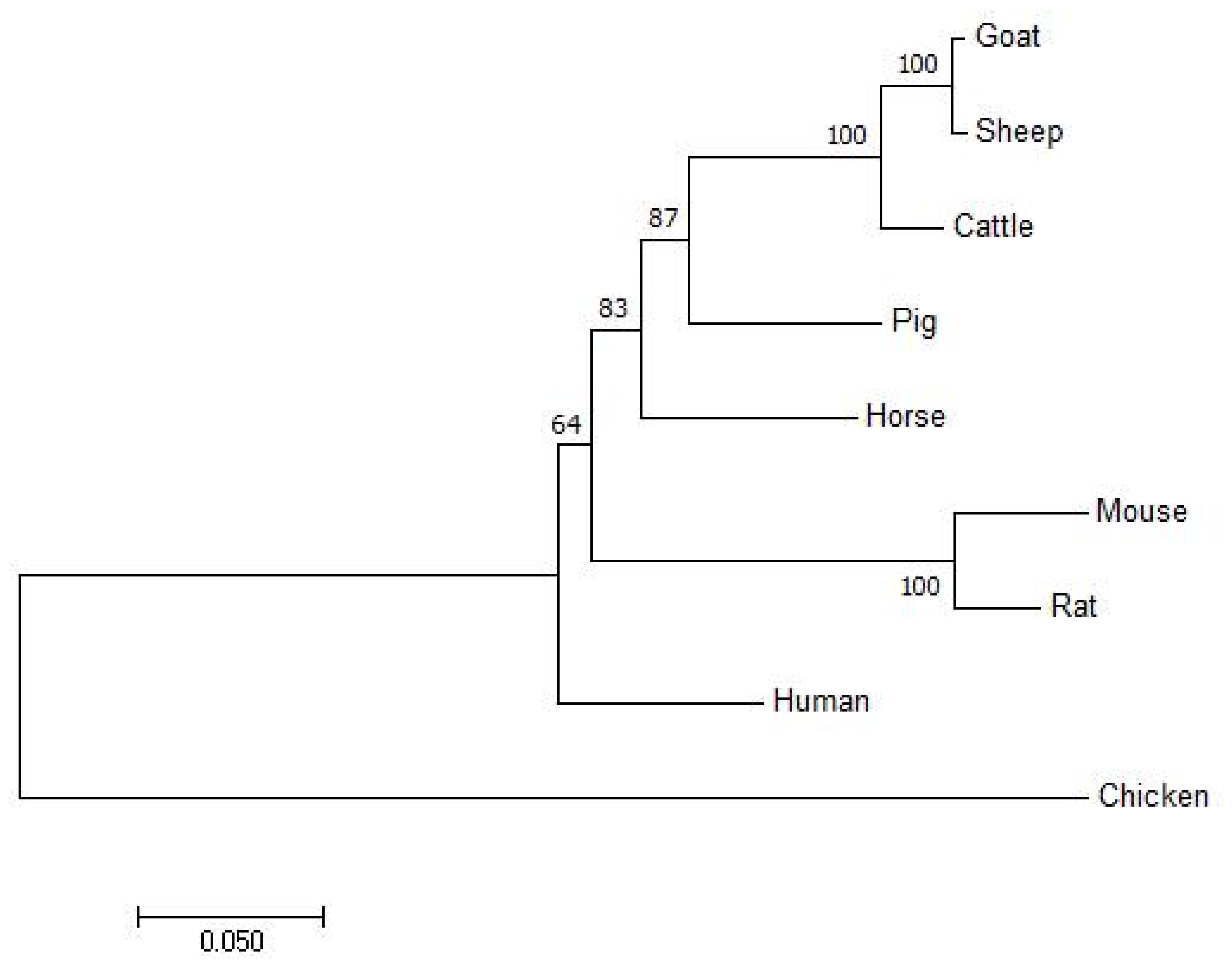
3.1. Species Homology, Phylogenetic Tree, and Conservative Estimation
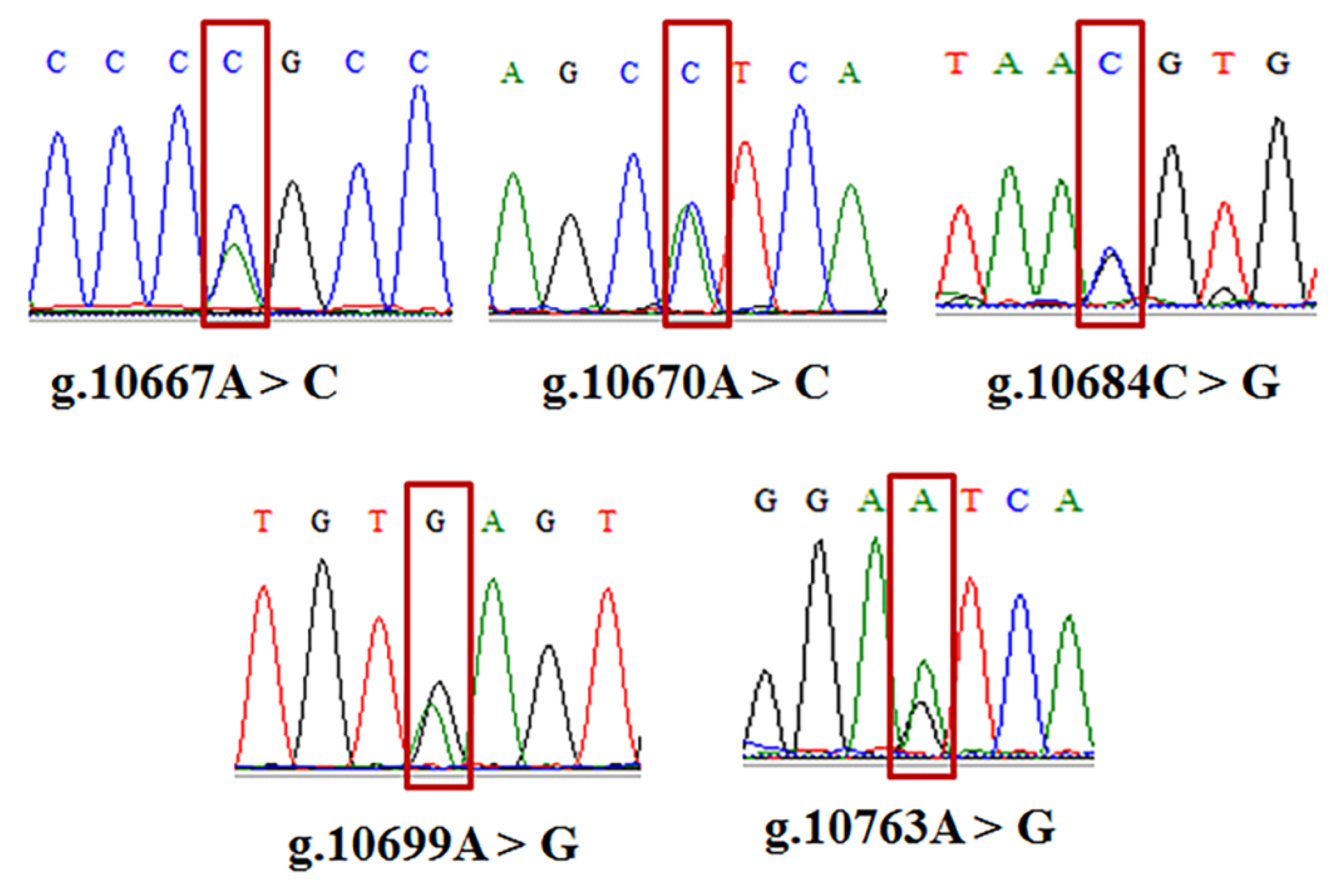
3.2. Identification of SNPs in GHRHR
3.3. Genetic Diversity, and Hardy Weinberg Equilibrium
3.4. Linkage Disequilibrium, and Haplotype Analysis
3.5. Association Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- China National Commission of Animal Genetic Resources. Animal Genetic Research in China, Bovines; Chinese Agricultural Press: Beijing, China, 2010; pp. 79–82. [Google Scholar]
- Cheng, M.; Shi, G.L.; Liu, S.Z.; Zhang, Y.H.; Liu, H.Y. Analysis of genetic variation and phylogeny on mtDNA D-loop regions in Dabieshan cattle. J. Anhui Agric. Univ. 2018, 45, 72–77. [Google Scholar]
- Li, F.Y.; Xia, X.T.; Jia, Y.T.; Dang, R.H.; Chen, H.; Lei, C.Z. Y-SNPs and Y-STRs Genetic Diversity and Paternal Origin of Dabieshan Cattle. China Cattle Sci. 2018, 44, 4–6. [Google Scholar]
- Mayo, K.E. Molecular cloning and expression of a pituitary-specific receptor for growth hormone-releasing hormone. Mol. Endocrinol. 1992, 6, 1734–1744. [Google Scholar] [PubMed]
- Lin-Su, K.; Wajnrajch, M.P. Growth hormone releasing hormone (GHRH) and the GHRH receptor. Rev. Endocr. Metab. Disord. 2002, 3, 313–323. [Google Scholar] [CrossRef]
- Salvatori, R.; Fan, X.; Mullis, P.E.; Haile, A.; Levine, M.A. Decreased expression of the GHRH receptor gene due to a mutation in a Pit-1 binding site. Mol. Endocrinol. 2002, 16, 450–458. [Google Scholar] [CrossRef]
- Kamijo, T.; Hayashi, Y.; Seo, H.; Yamamoto, M.; Ogawa, M.; Choski, C.S.; Sawant, N.J.; Colaco, M.P.; Desai, M.P. A nonsense mutation (E72X) in growth hormone releasing hormone receptor (GHRHR) gene is the major cause of familial isolated growth hormone deficiency in Western region of India: Founder effect suggested by analysis of dinucleotide repeat polymorphism close to GHRHR gene. Growth Horm. IGF Res. 2004, 14, 394–401. [Google Scholar]
- Hilal, L.; Hajaji, Y.; Vie-Luton, M.P.; Ajaltouni, Z.; Benazzouz, B.; Chana, M.; Chraibi, A.; Kadiri, A.; Amselem, S.; Sobrier, M.L. Unusual phenotypic features in a patient with a novel splice mutation in the GHRHR gene. Mol. Med. 2008, 14, 286–292. [Google Scholar] [CrossRef]
- Carakushansky, M.; Whatmore, A.J.; Clayton, P.E.; Shalet, S.M.; Gleeson, H.K.; Price, D.A.; Levine, M.A.; Salvatori, R. A new missense mutation in the growth hormone-releasing hormone receptor gene in familial isolated GH deficiency. Eur. J. Endocrinol. 2003, 148, 25–30. [Google Scholar] [CrossRef][Green Version]
- Inoue, H.; Kangawa, N.; Kinouchi, A.; Sakamoto, Y.; Kimura, C.; Horikawa, R.; Shigematsu, Y.; Itakura, M.; Ogata, T.; Fujieda, K.; et al. Identification and functional analysis of novel human growth hormone-releasing hormone receptor (GHRHR) gene mutations in Japanese subjects with short stature. Clin. Endocrinol. 2011, 74, 223–233. [Google Scholar] [CrossRef]
- Godfrey, P.; Rahal, J.O.; Beamer, W.G.; Copeland, N.G.; Jenkins, N.A.; Mayo, K.E. GHRH receptor of little mice contains a missense mutation in the extracellular domain that disrupts receptor function. Nat. Genet. 1993, 4, 227–232. [Google Scholar] [CrossRef]
- Wu, M.; Zhao, H.; Tang, X.; Li, Q.; Yi, X.; Liu, S.; Sun, X. Novel InDels of GHR, GHRH, GHRHR and Their Association with Growth Traits in Seven Chinese Sheep Breeds. Animals 2020, 10, 1883. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lan, X.Y.; Qu, Y.J.; Li, Z.J.; Chen, Z.Q.; Lei, C.Z.; Fang, X.T.; Chen, H. Effects of genetic variability of the dairy goat growth hormone releasing hormone receptor (GHRHR) gene on growth traits. Mol. Biol. Rep. 2011, 38, 539–544. [Google Scholar] [CrossRef]
- Zhang, C.F.; Chen, H.; Zhang, Z.Y.; Zhang, L.Z.; Yang, D.Y.; Qu, Y.J.; Hua, L.S.; Zhang, B.; Hu, S.R. A 5′UTR SNP of GHRHR locus is associated with body weight and average daily gain in Chinese cattle. Mol. Biol. Rep. 2012, 39, 10469–10473. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol.Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef]
- Wang, G.Q.; Zhang, S.; Wei, S.J.; Zhang, Y.R.; Li, Y.K.; Fu, C.Z.; Zhao, C.P.; Zan, L.S. Novel polymorphisms of SIX4 gene and their association with body measurement traits in Qinchuan cattle. Gene 2014, 539, 107–110. [Google Scholar] [CrossRef]
- Yang, W.C.; Wang, Y.N.; Fu, C.Z.; Zan, L.S. Association study and expression analysis of MTNR1A as a candidate gene for body measurement and meat quality traits in Qinchuan cattle. Gene 2015, 570, 199–204. [Google Scholar] [CrossRef]
- Chakraborty, R.; Nei, M. Bottleneck Effects on Average Heterozygosity and Genetic Distance with the Stepwise Mutation Model. Evolution 1977, 31, 347–356. [Google Scholar] [CrossRef]
- Slatkin, M. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef]
- Zhao, C.P.; Raza, S.H.A.; Khan, R.; Sabek, A.; Khan, S.; Ullah, I.; Memon, S.; El-Aziz, A.H.A.; Shah, M.A.; Shijun, L.; et al. Genetic variants in MYF5 affected growth traits and beef quality traits in Chinese Qinchuan cattle. Genomics 2020, 112, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.P.; Jin, H.; Xu, L.; Jia, Y.T. Genetic variants of the growth differentiation factor 8 affect body conformation traits in Chinese Dabieshan cattle. Anim. Biosci. 2022, 35, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Khan, R.; Schreurs, N.M.; Guo, H.F.; Gui, L.S.; Mei, C.G.; Zan, L.S. Expression of the bovine KLF6 gene polymorphisms and their association with carcass and body measures in Qinchuan cattle (Bos taurus). Genomics 2020, 112, 423–431. [Google Scholar] [CrossRef]
- Liang, W.C.; Ren, J.L.; Yu, Q.X.; Li, J.; Ng, T.K.; Chu, W.K.; Qin, Y.J.; Chu, K.O.; Schally, A.V.; Pang, C.P.; et al. Signaling mechanisms of growth hormone-releasing hormone receptor in LPS-induced acute ocular inflammation. Proc. Natl. Acad. Sci. USA 2020, 117, 6067–6074. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.K.; Law, K.S.; Chan, S.O.; Yam, J.C.; Chen, L.J.; Zhang, H.; Cheung, H.S.; Block, N.L.; Schally, A.V.; Pang, C.P. Antagonists of growth hormone-releasing hormone receptor induce apoptosis specifically in retinoblastoma cells. Proc. Natl. Acad. Sci. USA 2016, 113, 14396–14401. [Google Scholar] [CrossRef]
- Xu, Y.; Cai, H.F.; Zhou, Y.; Shi, T.; Lan, X.Y.; Zhang, C.L.; Lei, C.Z.; Jia, Y.T.; Chen, H. SNP and haplotype analysis of paired box 3 (PAX3) gene provide evidence for association with growth traits in Chinese cattle. Mol. Biol.Rep. 2014, 41, 4295–4303. [Google Scholar] [CrossRef]
- Wu, S.; Ning, Y.; Raza, S.H.A.; Zhang, C.T.; Zhang, L.; Cheng, G.; Wang, H.B.; Schreurs, N.; Zan, L.S. Genetic variants and haplotype combination in the bovine CRTC3 affected conformation traits in two Chinese native cattle breeds (Bos taurus). Genomics 2019, 111, 1736–1744. [Google Scholar] [CrossRef]
- Wen, Y.F.; Zheng, L.; Niu, H.; Zhang, G.L.; Zhang, G.M.; Ma, Y.L.; Tian, Y.R.; Liu, Y.R.; Yang, P.; Yang, D.Y.; et al. Exploring genotype-phenotype relationships of the CRABP2 gene on growth traits in beef cattle. Anim. Biotechnol. 2020, 31, 42–51. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Jiang, E.; Yan, H.; Zhu, H.; Chen, H.; Liu, J.; Qu, L.; Pan, C.; Lan, X.Y. InDels within caprine IGF2BP1 intron 2 and the 3′-untranslated regions are associated with goat growth traits. Anim. Genet. 2020, 51, 117–121. [Google Scholar] [CrossRef]
- Greenwood, T.A.; Kelsoe, J.R. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics 2003, 82, 511–520. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence | Tm (°C) | Position | Segment Length |
|---|---|---|---|---|
| GHRHR-F1 | 5′–GGAAAGGATGGGATTGTAGT–3′ | 60 | Intron 1 | 498 bp |
| GHRHR-R1 | 5′–CGGCAAAAGCATAGTGGGTA–3′ | |||
| GHRHR-F2 | 5′–TCCTGTCCCTCAAAGACCTG–3′ | 60 | Intron 9, exon 10, intron 10 | 612 bp |
| GHRHR-R2 | 5′–CCCTCTCTCAAATGCTCTGG–3′ |
| Gene | Number | Exon Size(bp) | Intron Size (bp) | 5′Splice Donor | 3′Splice Acceptor |
|---|---|---|---|---|---|
| GHRHR | 1 | 189 | 3892 | CGATCgtgag | cacagGTCCT |
| 2 | 103 | 128 | CTTGGgtacg | tctagGCTGC | |
| 3 | 108 | 746 | GCCAGgtgag | cctagGGGCT | |
| 4 | 98 | 1150 | AGGAGgtgag | cccagAAATC | |
| 5 | 98 | 512 | CTCAGgtttg | tccagGAGGC | |
| 6 | 133 | 1410 | CCACTgtaac | cacagGTCCT | |
| 7 | 154 | 300 | CTGGGgtgag | cacagGGCTT | |
| 8 | 61 | 569 | GTTGCgtgag | ttcagGTGCT | |
| 9 | 70 | 865 | TTGGGgtcag | tccagGTGAA | |
| 10 | 92 | 456 | TACTGgtaac | tgcagGCGTC | |
| 11 | 130 | 713 | TCCAGgtgag | cacagGGCTT | |
| 12 | 42 | 2852 | AAGAGgtatg | tctagGTGAC | |
| 13 | 1140 | – | – | – |
| SNP | Genotypic Frequency | Allelic Frequency | HWE | Diversity Parameter | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | A1 | A2 | χ2 | p-Value | Ho | He | Ne | PIC | |
| g.2052C > G | CC | GC | GG | C | G | ||||||
| 0.28 | 0.54 | 0.18 | 0.55 | 0.45 | 2.16 | 0.339 | 0.505 | 0.495 | 1.981 | 0.372 | |
| g.10667A > C | AA | AC | CC | A | C | ||||||
| 0.48 | 0.40 | 0.12 | 0.68 | 0.32 | 1.38 | 0.502 | 0.567 | 0.433 | 1.764 | 0.339 | |
| g.10670A > C | AA | AC | CC | A | C | ||||||
| 0.26 | 0.53 | 0.21 | 0.52 | 0.48 | 1.38 | 0.502 | 0.501 | 0.499 | 2.000 | 0.375 | |
| g.10684C > G | CC | GC | GG | C | G | ||||||
| 0.24 | 0.16 | 0.60 | 0.40 | 0.60 | 1.10 | 0.576 | 0.519 | 0.491 | 1.929 | 0.366 | |
| g.10699A > G | AA | AG | GG | A | G | ||||||
| 0.10 | 0.37 | 0.53 | 0.29 | 0.71 | 3.86 | 0.145 | 0.586 | 0.414 | 1.707 | 0.328 | |
| g.10763A>G | AA | AG | GG | A | G | ||||||
| 0.31 | 0.11 | 0.58 | 0.31 | 0.69 | 9.51 | 0.008 | 0.574 | 0.426 | 1.742 | 0.335 | |
| Item | g.2052C > G | g.10667A > C | g.10670A > C | g.10684C > G | g.10699A > G | g.10763A > G |
|---|---|---|---|---|---|---|
| g.2052C > G | r2 = 0.025 | r2 = 0.046 | r2 = 0.033 | r2 = 0.012 | r2 = 0.033 | |
| g.10667A > C | D’ = 0.278 | r2 = 0.138 | r2 = 0.124 | r2 = 0.729 | r2 = 0.121 | |
| g.10670A > C | D’ = 0.349 | D’ = 1.000 | r2 = 0.836 | r2 = 0.093 | r2 = 0.801 | |
| g.10684C > G | D’ = 0.255 | D’ = 0.870 | D’ = 0.992 | r2 = 0.097 | r2 = 0.919 | |
| g.10699A > G | D’ = 0.216 | D’ = 0.919 | D’ = 0.883 | D’ = 0.827 | r2 = 0.098 | |
| g.10763A > G | D’ = 0.257 | D’ = 0.849 | D’ = 0.992 | D’ = 0.979 | D’ = 0.825 |
| Haplotype | g.2052C > G | g.10667A > C | g.10670A > C | g.10684C > G | g.5148A > C | g.10699A > G | Frequency |
|---|---|---|---|---|---|---|---|
| Hap1 | C | A | A | C | G | A | 36.10% |
| Hap2 | G | A | C | G | G | G | 13.40% |
| Hap3 | C | C | C | G | A | G | 12.30% |
| Hap4 | G | A | A | C | G | A | 7.60% |
| Hap5 | G | C | C | G | A | G | 6.50% |
| Hap6 | C | A | C | G | G | G | 6.20% |
| SNP | Genotype | Traits (Mean ± SE) | ||||||
|---|---|---|---|---|---|---|---|---|
| BL (cm) | WH (cm) | HH (cm) | HG (cm) | AGR (cm) | HW (cm) | PBW (cm) | ||
| g.2052C > G | CC | 127.76 ± 2.04 | 112.59 ± 1.57 a | 110.58 ± 1.59 a | 148.02 ± 1.61 | 174.84 ± 1.40 a | 31.95 ± 0.41 | 17.45 ± 0.55 |
| CG | 124.93 ± 1.34 | 109.57 ± 1.69 b | 109.81 ± 1.50 ab | 146.39 ± 1.13 | 172.90 ± 1.45 ab | 30.41 ± 0.54 | 16.56 ± 0.45 | |
| GG | 123.60 ± 1.36 | 108.42 ± 1.03 b | 107.74 ± 0.93 b | 145.17 ± 1.62 | 168.64 ± 2.02 b | 30.90 ± 0.44 | 16.83 ± 0.66 | |
| p-value | 0.226 | 0.031 | 0.047 | 0.548 | 0.026 | 0.111 | 0.079 | |
| g.10667A > C | AA | 125.38 ± 1.67 | 110.36 ± 1.66 | 109.18 ± 1.58 | 150.96 ± 1.33 | 173.42 ± 1.74 | 33.38 ± 0.53 | 16.02 ± 0.29 a |
| AC | 125.41 ± 1.81 | 110.12 ± 1.54 | 109.19 ± 1.48 | 149.90 ± 1.23 | 175.07 ± 1.34 | 33.65 ± 0.48 | 17.35 ± 0.29 b | |
| CC | 125.87 ± 1.08 | 110.80 ± 1.65 | 109.21 ± 0.86 | 152.77 ± 1.33 | 172.54 ± 1.85 | 32.96 ± 0.55 | 16.15 ± 0.27 ab | |
| p-value | 0.954 | 0.869 | 0.999 | 0.636 | 0.770 | 0.825 | 0.039 | |
| g.10670A > C | AA | 124.53 ± 1.27 | 109.92 ± 1.39 | 109.16 ± 1.38 | 147.96 ± 1.22 | 171.43 ± 1.97 | 33.65 ± 0.63 | 15.41 ± 0.37 a |
| AC | 124.86 ± 1.87 | 109.34 ± 1.57 | 109.03 ± 0.96 | 150.22 ± 1.57 | 173.65 ± 1.44 | 33.28 ± 0.49 | 17.07 ± 0.29 b | |
| CC | 125.99 ± 0.97 | 110.88 ± 1.60 | 109.23 ± 1.46 | 152.34 ± 0.96 | 174.26 ± 1.66 | 33.23 ± 0.49 | 16.31 ± 0.28 ab | |
| p-value | 0.540 | 0.252 | 0.976 | 0.159 | 0.486 | 0.861 | 0.010 | |
| g.10684C > G | CC | 124.53 ± 1.17 | 107.12 ± 1.16 b | 109.46 ± 0.81 | 146.40 ± 1.74 | 171.98 ± 1.85 | 33.07 ± 0.60 | 15.81 ± 0.48 b |
| CG | 124.90 ± 0.89 | 110.45 ± 1.74 a | 108.00 ± 1.52 | 149.96 ± 1.41 | 172.33 ± 1.47 | 33.28 ± 0.51 | 16.29 ± 0.28 ab | |
| GG | 126.03 ± 1.97 | 110.91 ± 1.62 a | 109.22 ± 1.46 | 152.48 ± 1.26 | 174.46 ± 1.67 | 34.61 ± 0.42 | 17.15 ± 0.35 a | |
| p-value | 0.492 | 0.012 | 0.501 | 0.083 | 0.480 | 0.415 | 0.105 | |
| g.10699A > G | AA | 126.63 ± 1.13 | 110.74 ± 1.72 | 108.85 ± 1.58 | 153.65 ± 1.37 | 173.05 ± 1.87 | 32.80 ± 0.58 | 17.13 ± 0.28 a |
| AG | 123.56 ± 1.76 | 109.51 ± 1.51 | 109.20 ± 1.46 | 148.49 ± 1.95 | 172.62 ± 1.40 | 33.77 ± 0.42 | 15.05 ± 0.36 b | |
| GG | 125.62 ± 1.06 | 110.52 ± 1.66 | 109.25 ± 0.85 | 151.16 ± 1.33 | 173.76 ± 1.73 | 33.37 ± 0.54 | 16.10 ± 0.29 ab | |
| p-value | 0.347 | 0.596 | 0.920 | 0.279 | 0.905 | 0.713 | 0.046 | |
| g.10763A > G | AA | 124.31 ± 1.53 | 110.28 ± 1.75 a | 109.47 ± 1.72 | 150.01 ± 1.43 | 171.55 ± 1.78 | 33.13 ± 0.61 | 15.83 ± 0.29 |
| AG | 124.91 ± 0.95 | 106.79 ± 1.51 b | 108.00 ± 1.50 | 146.91 ± 1.25 | 173.29 ± 1.33 | 34.19 ± 0.37 | 16.90 ± 0.36 | |
| GG | 126.13 ± 1.46 | 110.95 ± 1.61 a | 109.26 ± 0.87 | 152.58 ± 1.28 | 174.87 ± 1.66 | 33.29 ± 0.54 | 16.30 ± 0.27 | |
| p-value | 0.492 | 0.049 | 0.441 | 0.174 | 0.070 | 0.280 | 0.185 | |
| Diplotypes | Frequency | BL (cm) | WH (cm) | HH (cm) | HG (cm) | AGR(cm) | HW (cm) | PBW (cm) |
|---|---|---|---|---|---|---|---|---|
| Hap 1/1 | 0.195 | 125.98 ± 1.39 | 110.62 ± 1.09 ab | 109.55 ± 0.76 ab | 152.57 ± 1.14 b | 173.65 ± 1.64 | 33.38 ± 0.34 b | 16.66 ± 0.47 |
| Hap 6/6 | 0.136 | 123.61 ± 1.21 | 107.91 ± 1.13 a | 106.09 ± 0.99 a | 149.08 ± 1.70 ab | 171.95 ± 1.76 | 32.54 ± 0.70 ab | 16.18 ± 0.81 |
| Hap 3/3 | 0.122 | 126.00 ± 1.10 | 110.03 ± 0.86 ab | 108.93 ± 0.84 ab | 154.06 ± 1.42 b | 173.43 ± 1.96 | 33.39 ± 0.62 b | 16.97 ± 0.44 |
| Hap 1/4 | 0.084 | 125.71 ± 1.25 | 112.29 ± 0.69 b | 111.68 ± 0.99 b | 149.00 ± 1.43 ab | 170.71 ± 1.66 | 33.50 ± 0.48 b | 16.93 ± 0.87 |
| Hap 2/2 | 0.174 | 125.26 ± 0.87 | 111.13 ± 0.92 b | 109.30 ± 0.93 ab | 153.54 ± 1.07 b | 174.62 ± 1.53 | 34.02 ± 0.48 b | 16.99 ± 0.58 |
| Hap 3/5 | 0.054 | 128.61 ± 1.04 | 114.15 ± 0.97 b | 112.31 ± 0.96 b | 155.74 ± 1.05 b | 174.08 ± 1.26 | 34.35 ± 0.59 b | 16.92 ± 0.83 |
| Hap 2/2 | 0.063 | 122.72 ± 0.98 | 110.17 ± 0.95 ab | 106.89 ± 0.81 a | 143.61 ± 1.14 a | 167.06 ± 1.40 | 30.89 ± 0.36 a | 15.88 ± 0.66 |
| Hap 2/5 | 0.066 | 124.42 ± 0.83 | 110.84 ± 0.97 ab | 109.67 ± 0.91 ab | 154.23 ± 0.92 b | 174.95 ± 1.43 | 34.00 ± 0.40 b | 16.97 ± 0.82 |
| p-value | 0.889 | 0.036 | 0.012 | 0.027 | 0.977 | 0.042 | 0.057 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Jin, H.; Xu, L.; Jia, Y. Polymorphisms of the Growth Hormone Releasing Hormone Receptor Gene Affect Body Conformation Traits in Chinese Dabieshan Cattle. Animals 2022, 12, 1601. https://doi.org/10.3390/ani12131601
Zhao S, Jin H, Xu L, Jia Y. Polymorphisms of the Growth Hormone Releasing Hormone Receptor Gene Affect Body Conformation Traits in Chinese Dabieshan Cattle. Animals. 2022; 12(13):1601. https://doi.org/10.3390/ani12131601
Chicago/Turabian StyleZhao, Shuanping, Hai Jin, Lei Xu, and Yutang Jia. 2022. "Polymorphisms of the Growth Hormone Releasing Hormone Receptor Gene Affect Body Conformation Traits in Chinese Dabieshan Cattle" Animals 12, no. 13: 1601. https://doi.org/10.3390/ani12131601
APA StyleZhao, S., Jin, H., Xu, L., & Jia, Y. (2022). Polymorphisms of the Growth Hormone Releasing Hormone Receptor Gene Affect Body Conformation Traits in Chinese Dabieshan Cattle. Animals, 12(13), 1601. https://doi.org/10.3390/ani12131601





