Genes Involved in Feed Efficiency Identified in a Meta-Analysis of Rumen Tissue from Two Populations of Beef Steers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cattle Populations
2.2. Sample Preparation
2.3. RNA-Sequencing
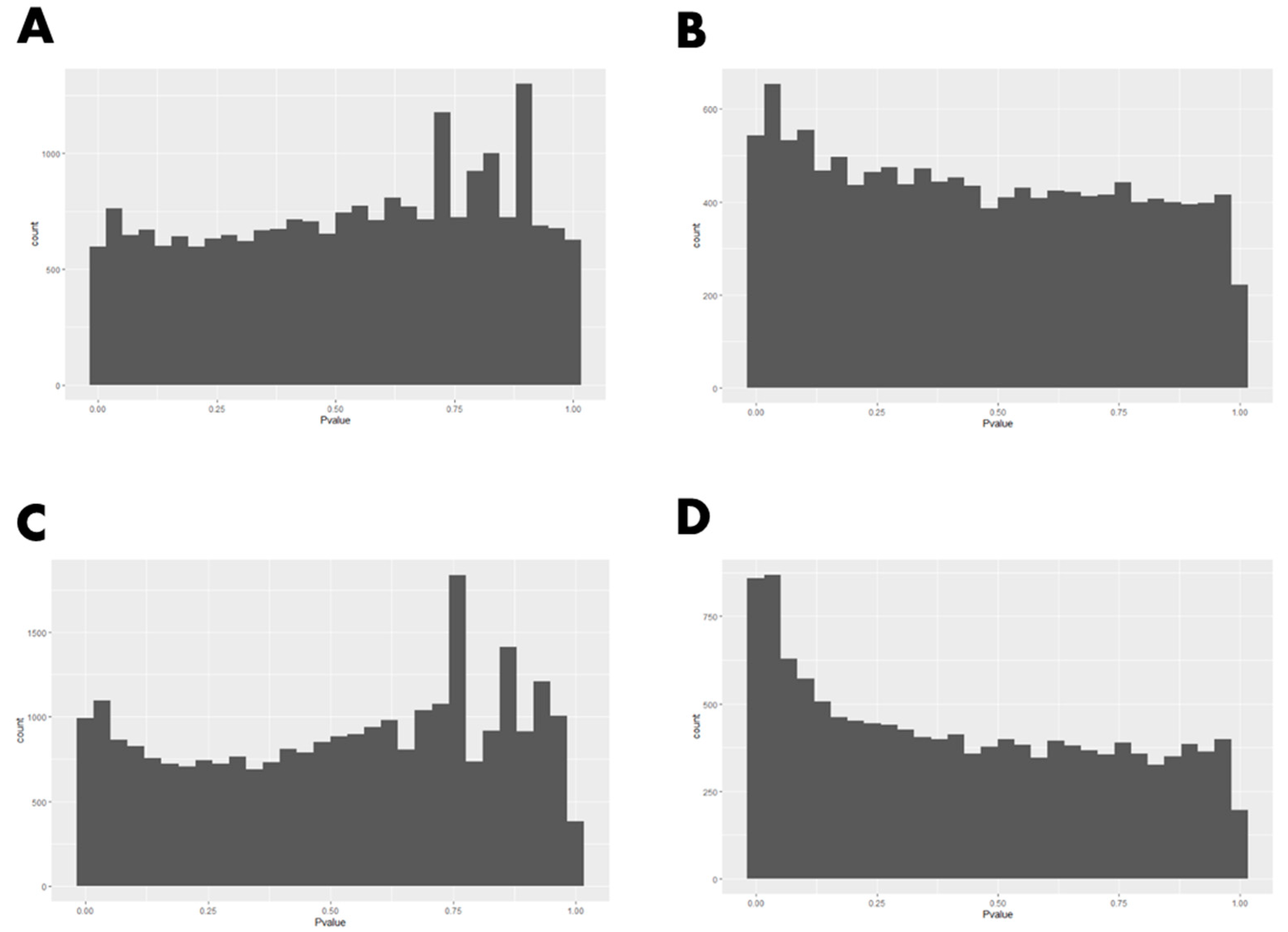
2.4. Data Analysis
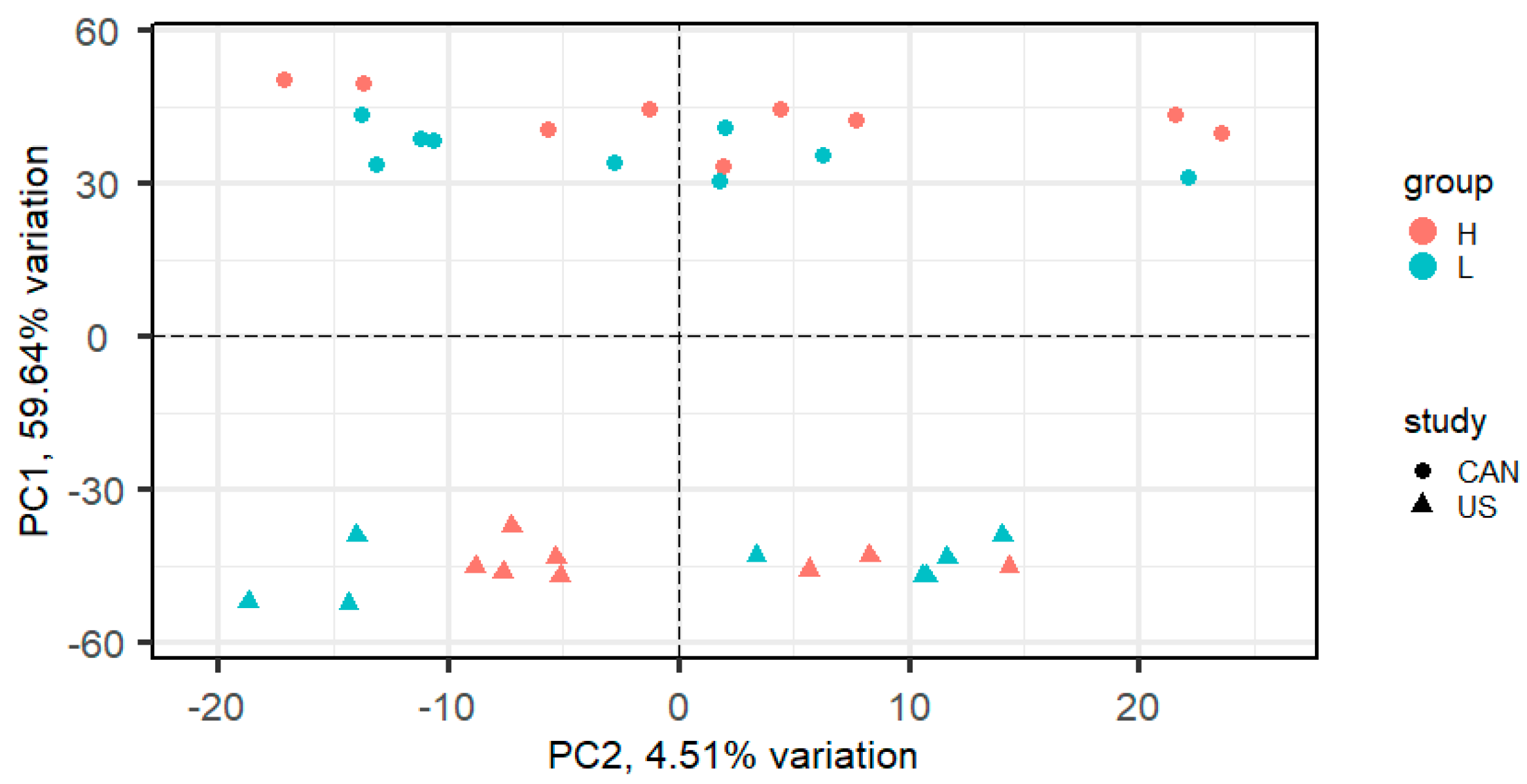
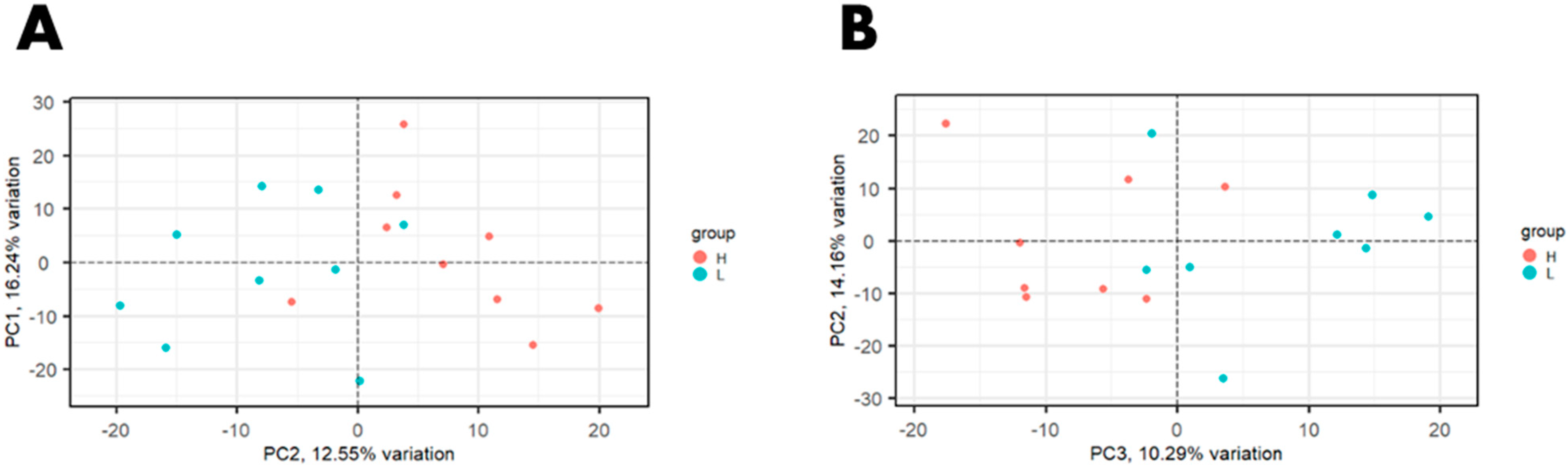
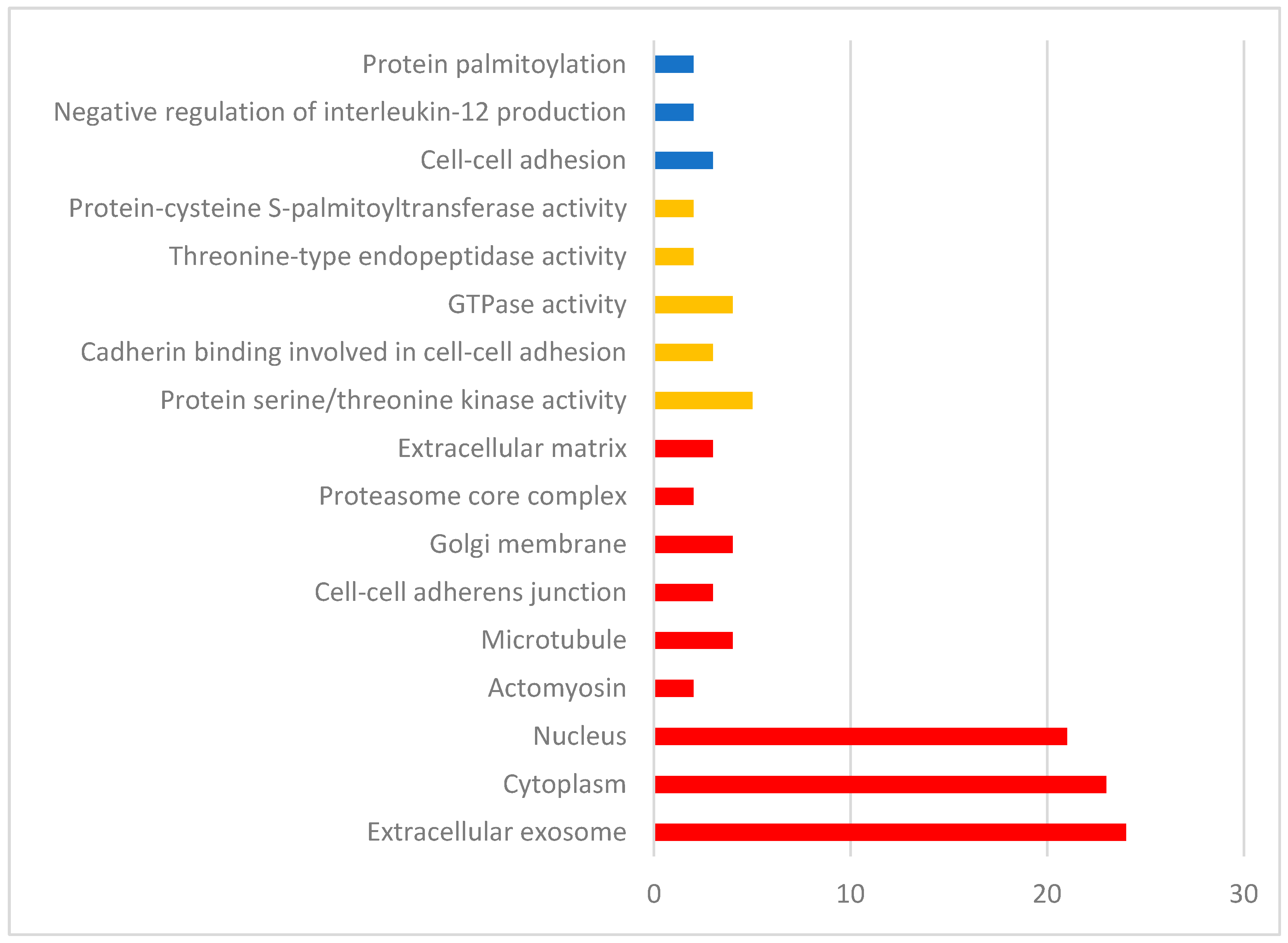
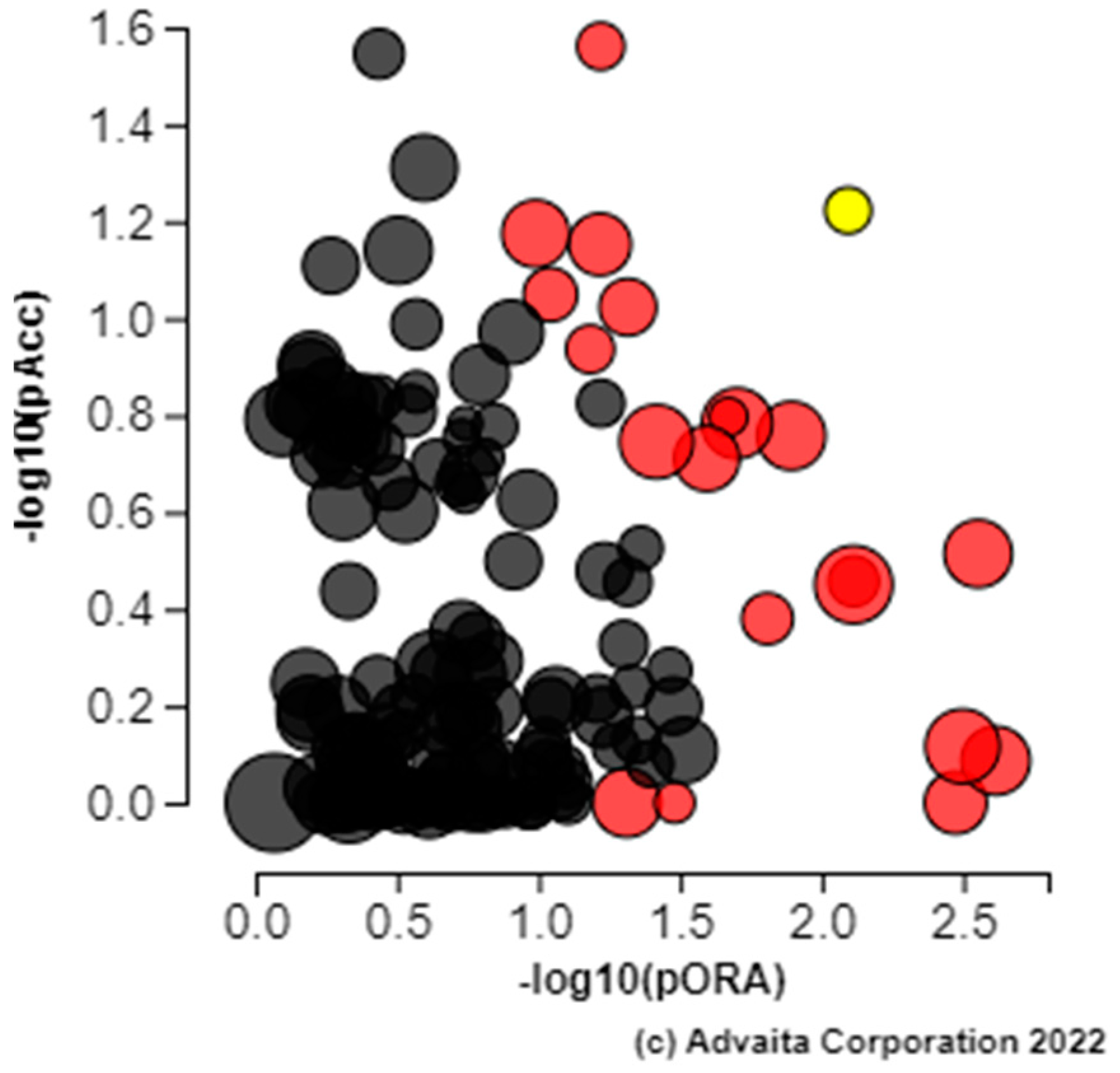
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S.; Guan, L.L. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C.; Berry, E.D.; Flythe, M.D.; Spangler, M.L.; Fernando, S.C. Rumen bacterial community structure impacts feed efficiency in beef cattle. J. Anim. Sci. 2018, 93, 1045–1058. [Google Scholar] [CrossRef]
- Delgado, B.; Bach, A.; Guasch, I.; González, C.; Elcoso, G.; Pryce, J.E.; Gonzalez-Recio, O. Whole rumen metagenome sequencing allows classifying and predicting feed efficiency and intake levels in cattle. Sci. Rep. 2019, 10, 2875. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.S.G.; Liang, G.; Chen, Y.; Stothard, P.; Guan, L.L. Transcriptome profiling of the rumen epithelium of beef cattle differing in residual feed intake. BMC Genom. 2016, 17, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kern, R.J.; Lindholm-Perry, A.K.; Freetly, H.C.; Snelling, W.M.; Kern, J.W.; Keele, J.W.; Miles, J.R.; Foote, A.P.; Oliver, W.T.; Kuehn, L.A.; et al. Transcriptome differences in the rumen of beef steers with variation in feed intake and gain. Gene 2016, 586, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Elolimy, A.A.; Abdelmegeid, M.K.; McCann, J.C.; Shike, D.W.; Loor, J.J. Residual feed intake in beef cattle and its associated with carcass traits, ruminal solid-fraction bacteria, and epithelium gene expression. J. Anim. Sci. Biotechnol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Freetly, H.C.; Oliver, W.T.; Rempel, L.A.; Keel, B.N. Genes associated with body weight gain and feed intake identified by meta-analysis of the mesenteric fat from crossbred beef steers. PLoS ONE 2020, 15, e0227154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keel, B.N.; Zarek, C.M.; Keele, J.W.; Kuehn, L.A.; Snelling, W.M.; Oliver, W.T.; Freetly, H.C.; Lindholm-Perry, A.K. RNA-Seq Meta-analysis identifies genes in skeletal muscle associated with gain and intake across a multi-season study of crossbred beef steers. BMC Genom. 2018, 19, 430. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, R.A. Statistical Methods for Research Workers; Oliver and Boyd: Edinburgh, UK, 1932. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Rau, A.; Marot, G.; Jaffrézic, F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinform. 2014, 15, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rau, A.; Gallopin, M.; Celeux, G.; Jaffrézic, F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics 2013, 29, 2146–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID gene functional classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elolimy, A.A.; Abdel-Hamied, E.; Hu, L.; McCann, J.C.; Shike, D.W.; Loor, J.J. Rapid Communication: Residual feed intake in beef cattle is associated with differences in protein turnover and nutrient transporters in ruminal epithelium. J. Anim. Sci. 2019, 97, 2181–2187. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X.G. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-H.; Shivdasani, R.A. Notch signaling in stomach epithelium stem cell homeostasis. J. Exp. Med. 2011, 208, 677–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obata, Y.; Takahashi, D.; Ebisawa, M.; Kakiguchi, K.; Yonemura, S.; Jinnohara, T.; Kanaya, T.; Fujimura, Y.; Ohmae, M.; Hase, K.; et al. Epithelial cell-intrinsic notch signaling plays an essential role in maintenance of gut immune homeostasis. J. Immunol. 2012, 188, 2427–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvidera, S.K.; Dickson, M.J.; Abuajamieh, M.; Snider, D.B.; Sanz Fernandez, M.V.; Johnson, J.S.; Keating, A.F.; Gorden, P.J.; Green, H.B.; Schoenberg, K.M.; et al. Intentionally induced intestinal barrier dysfunction causes inflammation, affects metabolism, and reduces productivity in lactating Holstein cows. J. Dairy Sci. 2017, 100, 4113–4127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, S.; Miglior, F.; Fonseca, P.A.S.; Gómez-Redondo, I.; Zeidan, J.; Suárez-Vega, A.; Schenkel, F.; Guan, L.L.; Waters, S.; Stothard, P.; et al. Identification of functional candidate variants and genes for feed efficiency in Holstein and Jersey cattle breeds using RNA-sequencing. J. Dairy Sci. 2021, 104, 1928–1950. [Google Scholar] [CrossRef] [PubMed]
- Gobé, C.; Elzaiat, M.; Meunier, N.; André, M.; Sellem, E.; Congar, P.; Jouneau, L.; Allais-Bonnet, A.; Naciri, I.; Passet, B.; et al. Dual role of DMXL2 in olfactory information transmission and the first wave of spermatogenesis. PLoS Genet. 2019, 15, e1007909. [Google Scholar] [CrossRef]
- Chen, Y.; Gondro, C.; Quinn, K.; Herd, R.M.; Parnell, P.F.; Vanselow, B. Global gene expression profiling reveals genes expressed differentially in cattle with high and low residual feed intake. Anim. Genet. 2011, 42, 475–490. [Google Scholar] [CrossRef]
- Grubbs, J.K.; Fritchen, A.N.; Huff-Lonergan, E.; Dekkers, J.C.; Gabler, N.K.; Lonergan, S.M. Divergent genetic selection for residual feed intake impacts mitochondria reactive oxygen species production in pigs. J. Anim. Sci. 2013, 91, 2133–2140. [Google Scholar] [CrossRef] [Green Version]
- Casal, A.; Garcia-Roche, M.; Nayajas, E.A.; Cassina, A.; Carriquiry, M. Differential hepatic oxidative status in steers with divergent residual feed intake phenotypes. Animal 2020, 14, 75–85. [Google Scholar] [CrossRef]
- Bottje, W.; Pumford, N.R.; Ojano-Dirain, C.; Iqbal, M.; Lassiter, K. Feed efficiency and mitochondrial function. Poult. Sci. 2006, 85, 8–14. [Google Scholar] [CrossRef]
- Del Bianco Benedeti, P.; Detmann, E.; Mantovani, H.C.; Bonilha, S.F.M.; Serão, N.V.L.; Lopes, D.R.G.; Silva, W.; Newbold, C.J.; Duarte, M.S. Nellore bulls (Bos taurus indicus) with high residual feed intake have increased the expression of genes involved in oxidative phosphorylation in rumen epithelium. Anim. Feed Sci. Technol. 2018, 235, 77–86. [Google Scholar] [CrossRef]
- Kolath, W.H.; Kerley, M.S.; Golden, J.W.; Keisler, D.H. The relationship between mitochondrial function and residual feed intake in Angus steers. J. Anim. Sci. 2006, 84, 861–865. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, P.A.; Carstens, G.E.; Michal, J.J.; Brennan, K.M.; Johnson, K.A.; Davis, M.E. Relationships between residual feed intake and hepatic mitochondrial function in growing beef cattle. J. Anim. Sci. 2014, 92, 3134–3141. [Google Scholar] [CrossRef]
- Snelling, W.M.; Allan, M.F.; Keele, J.W.; Kuehn, L.A.; Thallman, R.M.; Bennett, G.L.; Ferrell, C.L.; Jenkins, T.G.; Freetly, H.C.; Nielsen, M.K.; et al. Partial-genome evaluation of postweaning feed intake and efficiency of crossbred beef cattle. J. Anim. Sci. 2011, 89, 1731–1741. [Google Scholar] [PubMed] [Green Version]
- Lindholm-Perry, A.K.; Kern, R.J.; Kuehn, L.A.; Snelling, W.M.; Miles, J.R.; Oliver, W.T.; Freetly, H.C. Differences in transcript abundance of genes on BTA15 located within a region associated with gain in beef steers. Gene 2015, 572, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.W.; Shin, M.G.; Lee, S.; Kim, J.R.; Park, W.S.; Cho, K.H.; Meyer, T.; Heo, W.D. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell 2012, 47, 281–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoki, K.; Zhu, T.; Guan, K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [PubMed] [Green Version]
- Cruzen, S.M.; Harris, A.J.; Hollinger, K.; Punt, R.M.; Grubbs, J.K.; Selsby, J.T.; Dekkers, J.C.; Gabler, N.K.; Lonergan, S.M.; Huff-Lonergan, E. Evidence of decreased muscle protein turnover in gilts selected for low residual feed intake. J. Anim. Sci. 2013, 91, 4007–4016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene Symbol | PNominal | PFDR | Consistent LFC 1 |
|---|---|---|---|
| LOC789569 | 6.71 × 10−9 | 9.52 × 10−5 | N |
| LOC101904916 | 5.48 × 10−8 | 0.000389 | Y |
| TECR | 1.44 × 10−7 | 0.000680 | Y |
| ATP6AP1 | 2.96 × 10−7 | 0.000788 | N |
| PAMR1 | 3.11 × 10−7 | 0.000788 | N |
| EGLN3 | 7.51 × 10−7 | 0.00120 | N |
| LOC100848775 | 7.60 × 10−7 | 0.00120 | N |
| LYPD3 | 5.98 × 10−7 | 0.00120 | N |
| KAT2B | 2.46 × 10−6 | 0.00234 | Y |
| KLK13 | 2.22 × 10−6 | 0.00234 | N |
| PLP2 | 2.43 × 10−6 | 0.00234 | N |
| HTRA1 | 2.76 × 10−6 | 0.00245 | N |
| RHOG | 3.97 × 10−6 | 0.00313 | N |
| TUBA4A | 3.81 × 10−6 | 0.00313 | Y |
| CD52 | 7.06 × 10−6 | 0.00501 | Y |
| SH3BGRL3 | 7.81 × 10−6 | 0.00528 | N |
| SESN3 | 1.02 × 10−5 | 0.00636 | Y |
| ZDHHC5 | 1.07 × 10−5 | 0.00636 | Y |
| ZNF750 | 1.07 × 10−5 | 0.00636 | N |
| RPS15 | 1.85 × 10−5 | 0.0101 | Y |
| ODF2L | 2.70 × 10−5 | 0.0137 | Y |
| SH3GLB2 | 2.81 × 10−5 | 0.0138 | Y |
| HGS | 3.63 × 10−5 | 0.0172 | Y |
| MYL12A | 4.15 × 10−5 | 0.0190 | Y |
| ZDHHC3 | 4.57 × 10−5 | 0.0203 | N |
| ASB3 | 4.79 × 10−5 | 0.0205 | Y |
| MYADM | 4.90 × 10−5 | 0.0205 | Y |
| LOC104976804 | 5.56 × 10−5 | 0.0219 | Y |
| LYPD2 | 5.45 × 10−5 | 0.0219 | N |
| ASB2 | 5.90 × 10−5 | 0.0220 | N |
| CBX2 | 5.75 × 10−5 | 0.0220 | N |
| VARS | 6.67 × 10−5 | 0.0237 | Y |
| GLULP | 7.17 × 10−5 | 0.0242 | Y |
| RC3H1 | 7.02 × 10−5 | 0.0242 | Y |
| HSPB1 | 7.77 × 10−5 | 0.0251 | N |
| ZNF146 | 7.65 × 10−5 | 0.0251 | N |
| LY6G6C | 8.04 × 10−5 | 0.0254 | N |
| CYP1B1 | 9.02 × 10−5 | 0.0272 | N |
| PSMB5 | 9.26 × 10−5 | 0.0274 | Y |
| ALPK1 | 9.87 × 10−5 | 0.0277 | Y |
| DNM2 | 9.94 × 10−5 | 0.0277 | N |
| PSMB6 | 9.56 × 10−5 | 0.0277 | Y |
| B3GNT3 | 0.000119 | 0.0294 | N |
| C1QBP | 0.000117 | 0.0294 | Y |
| NBEAL1 | 0.000120 | 0.0294 | Y |
| SH3GL1 | 0.000110 | 0.0294 | Y |
| IL1RN | 0.000130 | 0.0312 | N |
| TUBB | 0.000135 | 0.0314 | Y |
| SLC35D1 | 0.000149 | 0.0328 | N |
| TMEM54 | 0.000152 | 0.0331 | N |
| LOC104971374 | 0.000171 | 0.0357 | N |
| CCDC66 | 0.000178 | 0.0367 | N |
| MAN2B1 | 0.000189 | 0.0382 | N |
| NDUFA9 | 0.000194 | 0.0382 | Y |
| CFL1 | 0.000197 | 0.0383 | Y |
| PIBF1 | 0.000199 | 0.0383 | N |
| C7H5orf46 | 0.000208 | 0.0384 | N |
| LOC100848030 | 0.000206 | 0.0384 | N |
| YPEL3 | 0.000204 | 0.0384 | N |
| MTERF2 | 0.000216 | 0.0392 | N |
| FRK | 0.000219 | 0.0394 | Y |
| ATR | 0.000239 | 0.0409 | N |
| REXO5 | 0.000238 | 0.0409 | N |
| RUVBL1 | 0.000251 | 0.0425 | Y |
| LOC104973218 | 0.000257 | 0.0425 | N |
| PRR5 | 0.000258 | 0.0425 | Y |
| DNAJB1 | 0.000265 | 0.0432 | N |
| MTAP | 0.000274 | 0.0438 | N |
| MAPK1 | 0.000278 | 0.0439 | Y |
| TMSB10 | 0.000298 | 0.0461 | N |
| UACA | 0.000298 | 0.0461 | Y |
| ARAF | 0.000305 | 0.0465 | N |
| DCUN1D4 | 0.000317 | 0.0465 | Y |
| GABARAP | 0.000315 | 0.0465 | N |
| MALL | 0.000318 | 0.0465 | N |
| RGS5 | 0.000316 | 0.0465 | Y |
| FAM107B | 0.000335 | 0.0476 | Y |
| LOC100139345 | 0.000345 | 0.0476 | N |
| PNPT1 | 0.000346 | 0.0476 | N |
| RWDD3 | 0.000334 | 0.0476 | N |
| SNX15 | 0.000341 | 0.0476 | N |
| ELF5 | 0.000354 | 0.0483 | Y |
| S100A11 | 0.000360 | 0.0486 | Y |
| Pathway | #DEG 1 | P | Genes |
|---|---|---|---|
| Renal Cell Carcinoma | 3 | 0.004 | ARAF, EGLN3, MAPK1 |
| Salmonella Infection | 6 | 0.007 | DNM2, MAPK1, MYL12A, RHOG, TUBA4A, TUBB |
| p53 Signaling Pathway | 2 | 0.012 | ATR, SESN3 |
| Phagosome | 4 | 0.012 | ATP6AP1, HGS, TUBA4A, TUBB |
| Prion Disease | 6 | 0.014 | MAPK1, NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Parkinson Disease | 5 | 0.016 | NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Alzheimer Disease | 5 | 0.017 | ARAF, MAPK1, NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Gap Junction | 3 | 0.019 | MAPK1, TUBA4A, TUBB |
| Pathways of Neurodegeneration—Multiple Diseases | 7 | 0.019 | ARAF, MAPK1, NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Huntington Disease | 5 | 0.022 | NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Bladder Cancer | 2 | 0.023 | ARAF, MAPK1, NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Axon Guidance | 3 | 0.028 | CFL1, MAPK1, MYL12A |
| Serotonergic Synapse | 2 | 0.029 | ARAF, MAPK1 |
| Regulation of Actin Cytoskeleton | 4 | 0.031 | ARAF, CFL1, MAPK1, MYL12A |
| Fc Gamma R-mediated Phagocytosis | 3 | 0.039 | CFL1, DNM2, MAPK1 |
| Human T-cell Leukemia Virus 1 Infection | 3 | 0.041 | ATR, KAT2B, MAPK1 |
| Amyotrophic Lateral Sclerosis | 5 | 0.041 | NDUFA9, PSMB5, PSMB6, TUBA4A, TUBB |
| Bacterial Invasion of Epithelial Cells | 2 | 0.045 | DNM2, RHOG |
| Parathyroid Hormone Synthesis, Secretion and Action | 2 | 0.047 | ARAF, MAPK1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindholm-Perry, A.K.; Meyer, A.M.; Kern-Lunbery, R.J.; Cunningham-Hollinger, H.C.; Funk, T.H.; Keel, B.N. Genes Involved in Feed Efficiency Identified in a Meta-Analysis of Rumen Tissue from Two Populations of Beef Steers. Animals 2022, 12, 1514. https://doi.org/10.3390/ani12121514
Lindholm-Perry AK, Meyer AM, Kern-Lunbery RJ, Cunningham-Hollinger HC, Funk TH, Keel BN. Genes Involved in Feed Efficiency Identified in a Meta-Analysis of Rumen Tissue from Two Populations of Beef Steers. Animals. 2022; 12(12):1514. https://doi.org/10.3390/ani12121514
Chicago/Turabian StyleLindholm-Perry, Amanda K., Allison M. Meyer, Rebecca J. Kern-Lunbery, Hannah C. Cunningham-Hollinger, Taran H. Funk, and Brittney N. Keel. 2022. "Genes Involved in Feed Efficiency Identified in a Meta-Analysis of Rumen Tissue from Two Populations of Beef Steers" Animals 12, no. 12: 1514. https://doi.org/10.3390/ani12121514
APA StyleLindholm-Perry, A. K., Meyer, A. M., Kern-Lunbery, R. J., Cunningham-Hollinger, H. C., Funk, T. H., & Keel, B. N. (2022). Genes Involved in Feed Efficiency Identified in a Meta-Analysis of Rumen Tissue from Two Populations of Beef Steers. Animals, 12(12), 1514. https://doi.org/10.3390/ani12121514






