Identification of the TSSK4 Alternative Spliceosomes and Analysis of the Function of the TSSK4 Protein in Yak (Bos grunniens)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Sample Collection
2.3. RNA Extraction and First-Strand cDNA Synthesis
2.4. Primer Design and Synthesis
2.5. PCR Amplification, Cloning and Sequencing of the TSSK4 Gene
2.6. Prokaryotic Expression and Purification of the TSSK4 Protein
2.7. GST Pull-Down Verifying the Interaction Protein of the TSSK4 Protein
2.8. Enzymolysis in the Protein Gel
2.9. Zip-Tip Desalting
2.10. LC–MS/MS Analysis
2.11. Data Analysis
3. Results
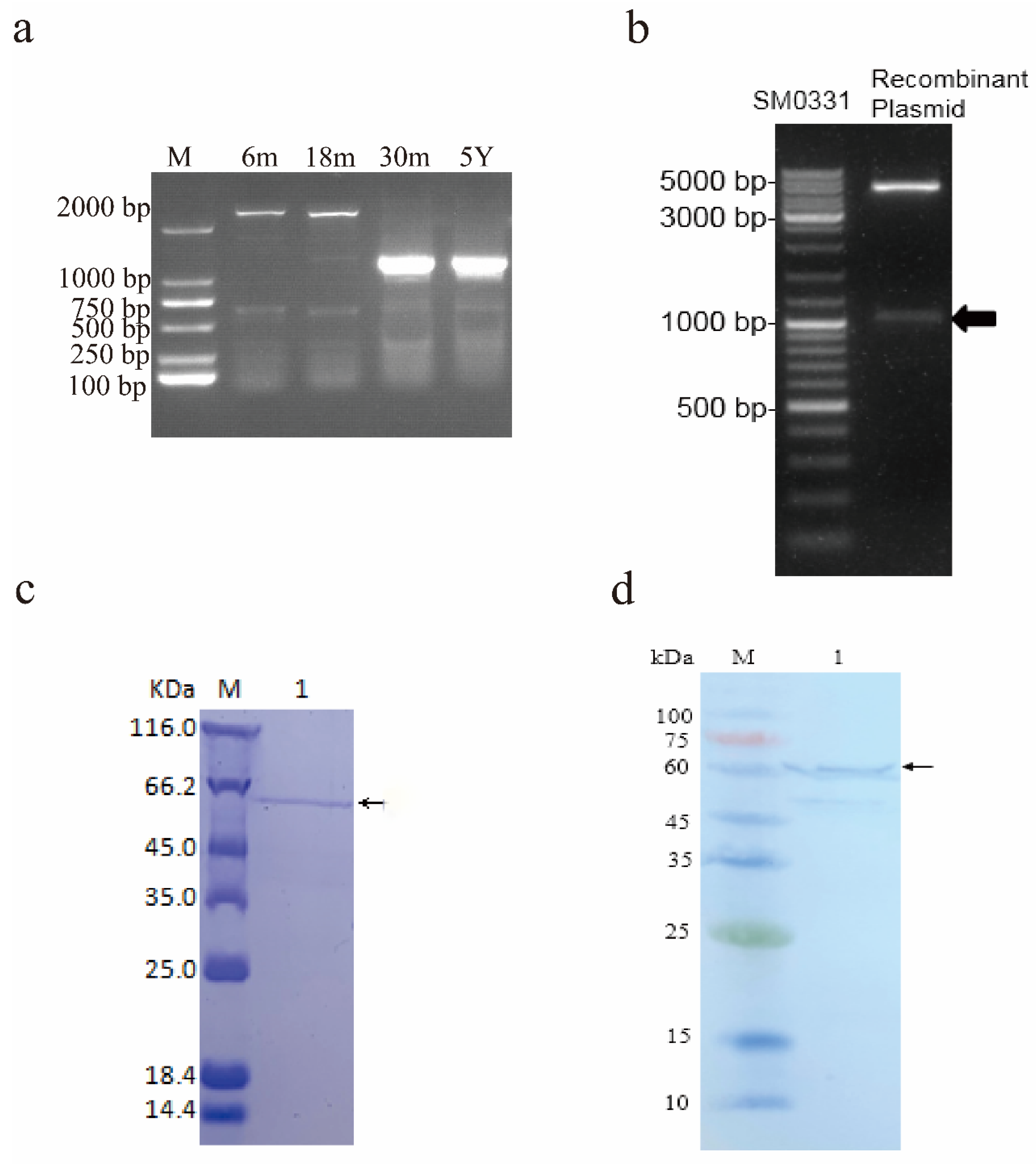
3.1. Identification and Subcellular Localization of the Alternative Spliceosomes of the Yak Tssk4
3.2. Prokaryotic Expression and Purification of the TSSK4 Protein
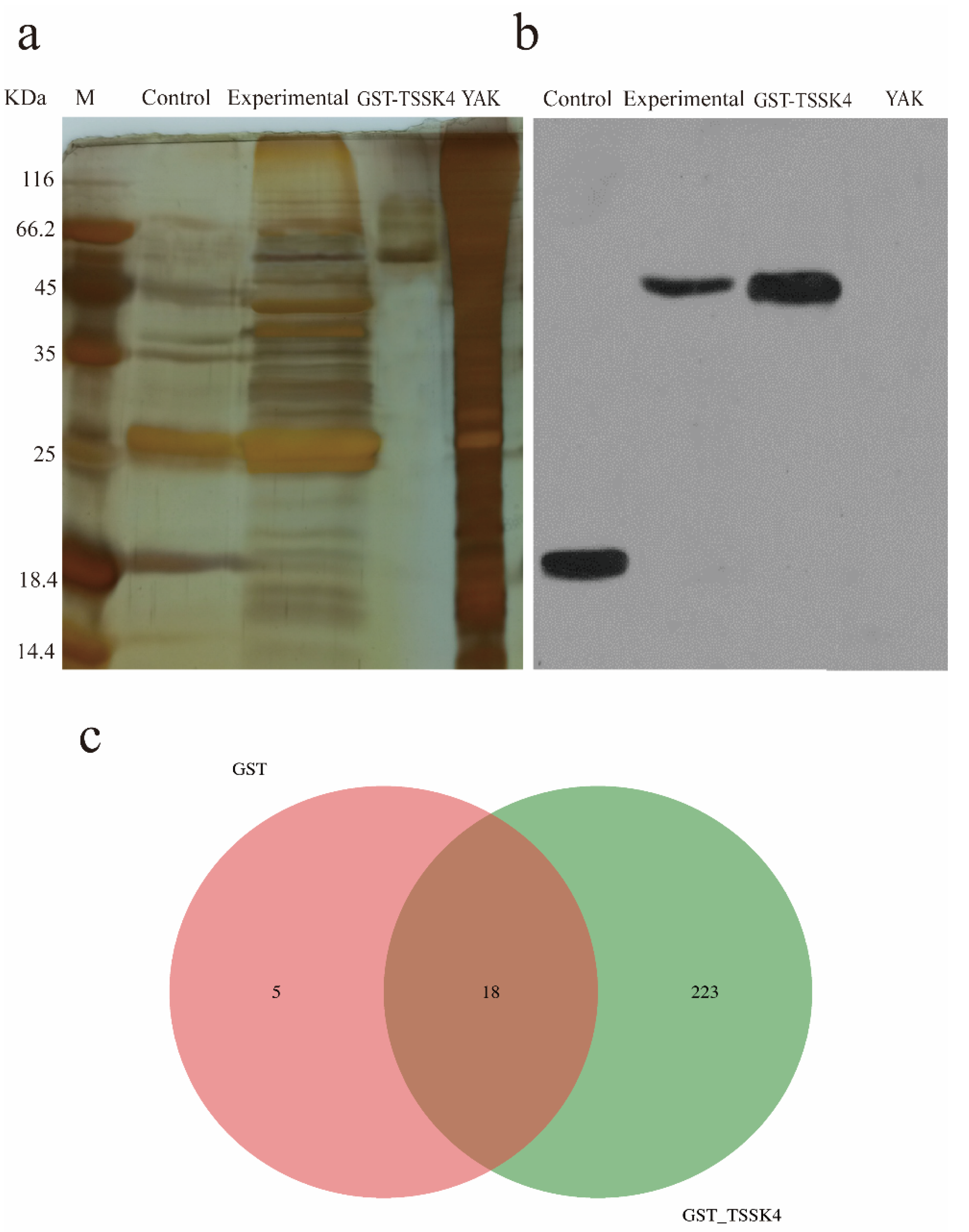
3.3. Identification of the TSSK4-Interacting Proteins by GST Pull-Down Combined with the LC–MS/MS Mass Spectrometry
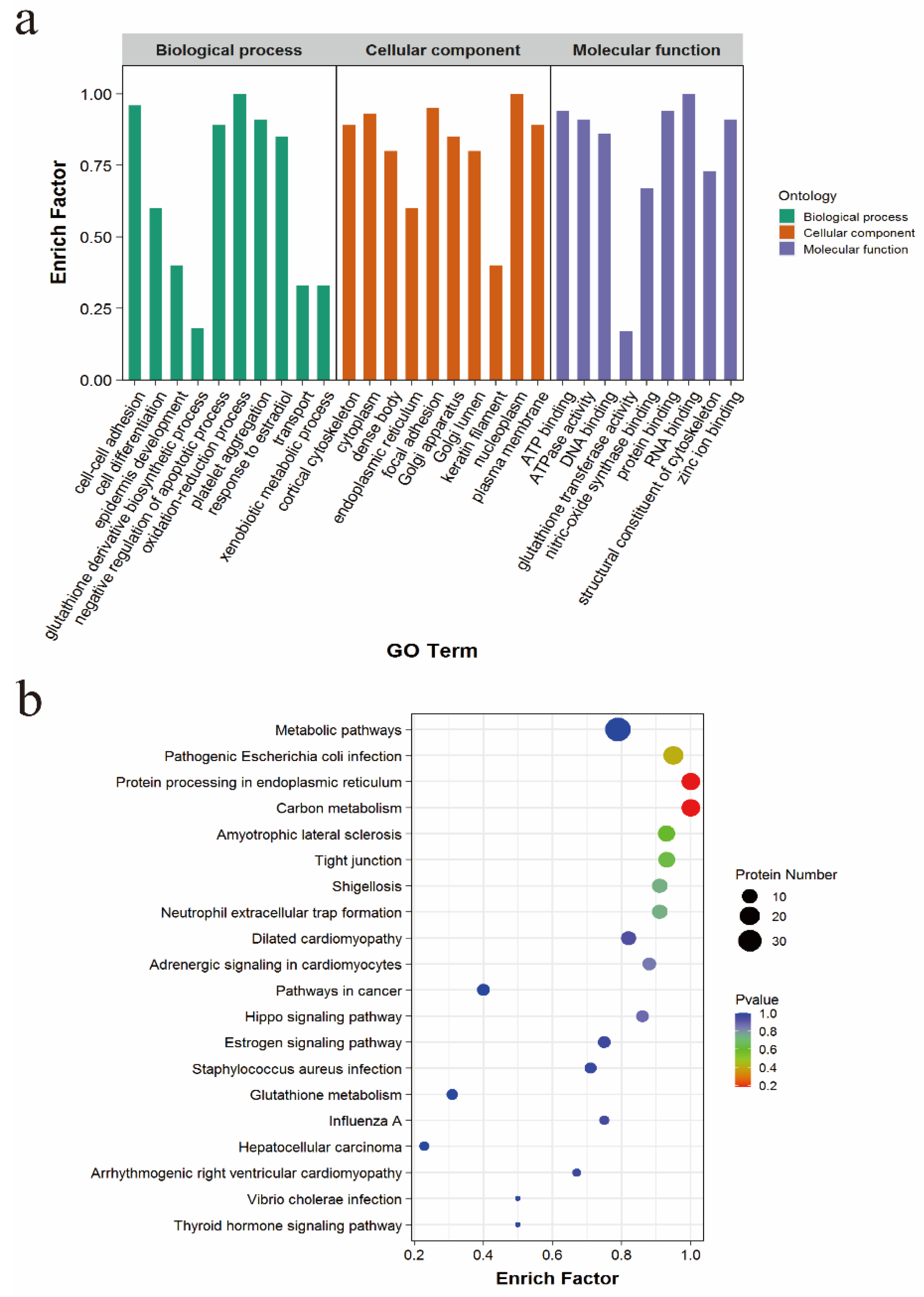
3.4. Enrichment Analysis of the TSSK4-Interacting Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, P.P.; Krishnan, G.; Doley, J.; Biswas, T.K.; Paul, V.; Chakravarty, P.; Deb, S.M.; Das, P.J. Identification and expression profiling of MSY genes of yak for bull fertility. J. Genet. 2019, 98, 41. [Google Scholar] [CrossRef] [PubMed]
- Hui, W.; Chai, Z.; Dan, H.; Ji, Q.; Zhong, J. A global analysis of CNVs in diverse yak populations using whole-genome resequencing. BMC Genom. 2019, 20, 61. [Google Scholar]
- Yang, C.; Ahmad, A.A.; Bao, P.J.; Guo, X.; Ding, X.Z. Increasing dietary energy level improves growth performance and lipid metabolism through up-regulating lipogenic gene expression in yak (Bos grunniens). Anim. Feed Sci. Technol. 2020, 263, 114455. [Google Scholar] [CrossRef]
- Wiener, G.; Han, J.; Long, R. The yak. Rap Publ. 2003, 44, 57–58. [Google Scholar]
- Wang, X.; Pei, J.; Bao, P.; Cao, M.; Guo, S.; Song, R.; Song, W.; Liang, C.; Yan, P.; Guo, X. Mitogenomic diversity and phylogeny analysis of yak (Bos grunniens). BMC Genom. 2021, 22, 325. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, M. Metabolomic profiles in yak mammary gland tissue during the lactation cycle. PLoS ONE 2019, 14, e0219220. [Google Scholar] [CrossRef]
- Jiang, M.; Lee, J.; Bionaz, M.; Deng, X.; Wang, Y. Evaluation of Suitable Internal Control Genes for RT-qPCR in Yak Mammary Tissue during the Lactation Cycle. PLoS ONE 2016, 11, e0147705. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, X.; Chu, M.; Liang, C.; Yan, P. Validation of Suitable Reference Genes for Gene Expression Studies on Yak Testis Development. Animals 2020, 10, 182. [Google Scholar] [CrossRef]
- Wang, P.; Huo, H.L.; Wang, S.Y.; Miao, Y.W.; Xiao, H. Cloning, sequence characterization, and expression patterns of members of the porcine TSSK family. Genet. Mol. Res. 2015, 14, 14908. [Google Scholar] [CrossRef]
- Wang, Z.; Cole, P.A. Catalytic Mechanisms and Regulation of Protein Kinases. Methods Enzymol. 2014, 548, 1–21. [Google Scholar]
- Wang, X.; Wei, Y.; Fu, G.; Li, H.; Saiyin, H.; Lin, G.; Wang, Z.; Chen, S.; Yu, L. Tssk4 is essential for maintaining the structural integrity of sperm flagellum. Mol. Hum. Reprod. 2015, 21, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Zhang, W.; Yang, Y.; Deng, Y.; Ma, Y.; Song, H.; Zhang, S. Mutation Screening and Association Study of the TSSK4 Gene in Chinese Infertile Men With Impaired Spermatogenesis. J. Androl. 2008, 29, 374–378. [Google Scholar] [CrossRef]
- Shang, P.; Baarends, W.M.; Hoogerbrugge, J.; Ooms, M.P.; Cappellen, W.V.; Jong, A.D.; Dohle, G.R.; Eenennaam, H.V.; Gossen, J.A.; Grootegoed, J.A. Functional transformation of the chromatoid body in mouse spermatids requires testis-specific serine/threonine kinases. J. Cell Sci. 2010, 123, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Jha, K.; Coleman, A.; Wong, L.; Salicioni, A.; Howcroft, E.; Johnson, G. Heat shock protein 90 functions to stabilize and activate the testis-specific serine/threonine kinases, a family of kinases essential for male fertility. J. Biol. Chem. 2013, 288, 16308–16320. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, H.; Fu, G.; Wang, Y.; Du, S.; Yu, L.; Wei, Y.; Chen, S. Testis-specific serine/threonine protein kinase 4 (Tssk4) phosphorylates Odf2 at Ser-76. Sci. Rep. 2016, 6, 22861. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, G.; Wei, Y.; Hexige, S.; Niu, Y.; Liu, L.; Yang, C.; Yu, L. TSSK5, a novel member of the testis-specific serine/threonine kinase family, phosphorylates CREB at Ser-133, and stimulates the CRE/CREB responsive pathway. Biochem. Biophys. Res. Commun. 2005, 333, 742–749. [Google Scholar] [CrossRef]
- Kueng, P.; Nikolova, Z.; Djonov, V.; Hemphill, A.; Rohrbach, V.; Boehlen, D.; Zuercher, G.; Andres, A.; Ziemiecki, A. A novel family of serine/threonine kinases participating in spermiogenesis. J. Cell Biol. 1997, 139, 1851–1859. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, X.; Fu, G.; Yu, L. Testis specific serine/threonine kinase 4 (Tssk4) maintains its kinase activity by phosphorylating itself at Thr-197. Mol. Biol. Rep. 2013, 40, 439–447. [Google Scholar] [CrossRef]
- Salicioni, A.M.; Gervasi, G.M.; Julian, S.; Tourzani, D.A.; Saman, N.; Caraballo, D.A.; Visconti, P.E. Testis-specific serine kinase protein family in male fertility and as targets for non-hormonal male contraception. Biol. Reprod. 2020, 103, 264–274. [Google Scholar] [CrossRef]
- Wang, X.; Wei, Y.; Fu, G.; Yu, L. Testis specific serine/threonine protein kinase 4 (TSSK4) leads to cell apoptosis relying on its kinase activity. J. Huazhong Univ. Sci. Technol. Med. Sci. 2015, 35, 235–240. [Google Scholar] [CrossRef]
- Sun, X.; Tian, Y.; Wang, J.; Sun, Z.; Zhu, Y. Genome-wide analysis reveals the association between alternative splicing and DNA methylation across human solid tumors. BMC Med. Genom. 2020, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Mthembu, N.N.; Zukile, M.; Rodney, H.; Zodwa, D. Abnormalities in alternative splicing of angiogenesis-related genes and their role in HIV-related cancers. HIV AIDS 2017, 9, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S. Alternative splicing, RNA-seq and drug discovery. Drug Discov. Today 2019, 24, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Bowler, E.; Oltean, S. Alternative Splicing in Angiogenesis. Int. J. Mol. Sci. 2019, 20, 2067. [Google Scholar] [CrossRef]
- Matlin, A.; Clark, F.; Smith, C. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef]
- Choi, N.; Liu, Y.; Oh, J.; Ha, J.; Zheng, X.; Shen, H. U2AF65-Dependent SF3B1 Function in SMN Alternative Splicing. Cells 2020, 9, 2647. [Google Scholar] [CrossRef]
- Du, J.X.; Zhu, G.Q.; Cai, J.L.; Wang, B.; Dai, Z. Splicing factors: Insights into their regulatory network in alternative splicing in cancer. Cancer Lett. 2020, 501, 83–104. [Google Scholar] [CrossRef]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Jin, L.; Chen, Y.; Crossman, D.K.; Datta, A.; Vu, T.; Mobley, J.A.; Basu, M.K.; Scarduzio, M.; Wang, H.; Chang, C. STRAP regulates alternative splicing fidelity during lineage commitment of mouse embryonic stem cells. Nat. Commun. 2020, 11, 5941. [Google Scholar] [CrossRef]
- Biamonti, G.; Catillo, M.; Pignataro, D.; Montecucco, A.; Ghigna, C. The alternative splicing side of cancer. Semin. Cell Dev. Biol. 2014, 32, 30–36. [Google Scholar] [CrossRef]
- Paronetto, M.P.; Passacantilli, I.; Sette, C. Alternative splicing and cell survival: From tissue homeostasis to disease. Cell Death Differ. 2016, 23, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Song, W.; Xie, D.; Li, Y. cDNA Cloning, Expression and Bioinformatical Analysis of Tssk Genes in Tree Shrews. Comput. Biol. Chem. 2021, 92, 107474. [Google Scholar] [CrossRef] [PubMed]
- Salicioni, A.M. Expression and localization of five members of the testis-specific serine kinase (Tssk) family in mouse and human sperm and testis. Mol. Hum. Reprod. 2011, 17, 42. [Google Scholar]
- Chen, Y.M.; Chen, C.; Riley, D.J.; Allred, D.C.; Lee, W.H. Aberrant Subcellular Localization of BRCA1 in Breast Cancer. Science 1995, 270, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.C.; Link, W. Protein localization in disease and therapy. J. Cell Sci. 2011, 124, 3381–3392. [Google Scholar] [CrossRef]
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An Actin-Dependent Step in Mitochondrial Fission Mediated by the ER-Associated Formin INF2. Science 2013, 339, 464. [Google Scholar] [CrossRef]
- Loveland, K.; Hayes, T.; Meinhardt, A.; Zlatic, K.; Parvinen, M.; de Kretser, D.; McFarlane, J. Microtubule-associated protein-2 in the rat testis: A novel site of expression. Biol. Reprod. 1996, 54, 896–904. [Google Scholar] [CrossRef][Green Version]
- Berthrong, M.; Goodwin, W.E.; Scott, W.W. Estrogen production by the testis. J. Clin. Endocrinol. Metab. 1949, 9, 579. [Google Scholar] [CrossRef]
- Stumpf, W.E.; Narbaitz, R.; Sar, M. Estrogen receptors in the fetal mouse. J. Steroid Biochem. 1980, 12, 55–64. [Google Scholar] [CrossRef]
- Walter, P.; Green, S.; Greene, G.; Krust, A.; Bornert, J.; Jeltsch, J.; Staub, A.; Jensen, E.; Scrace, G.; Waterfield, M. Cloning of the human estrogen receptor cDNA. Proc. Natl. Acad. Sci. USA 1985, 82, 7889–7893. [Google Scholar] [CrossRef]
- Greene, G.; Closs, L.; Fleming, H.; DeSombre, E.; Jensen, E. Antibodies to estrogen receptor: Immunochemical similarity of estrophilin from various mammalian species. Proc. Natl. Acad. Sci. USA 1977, 74, 3681–3685. [Google Scholar] [CrossRef] [PubMed]
- Cooke, P.; Young, P.; Hess, R.; Cunha, G. Estrogen receptor expression in developing epididymis, efferent ductules, and other male reproductive organs. Endocrinology 1991, 128, 2874–2879. [Google Scholar] [CrossRef] [PubMed]
- Rumi, M.; Dhakal, P.; Kubota, K.; Chakraborty, D.; Lei, T.; Larson, M.; Wolfe, M.; Roby, K.; Vivian, J.; Soares, M. Generation of Esr1-knockout rats using zinc finger nuclease-mediated genome editing. Endocrinology 2014, 155, 1991–1999. [Google Scholar] [CrossRef] [PubMed]
- Cooke, P.S.; Nanjappa, M.K.; Ko, C.M.; Prins, G.S.; Hess, R.A. Estrogens in Male Physiology. Physiol. Rev. 2017, 97, 995–1043. [Google Scholar] [CrossRef]
- Dumasia, K.; Kumar, A.; Deshpande, S.; Balasinor, N.H. Estrogen, through estrogen receptor 1, regulates histone modifications and chromatin remodeling during spermatogenesis in adult rats. Epigenetics 2017, 12, 953–963. [Google Scholar] [CrossRef]
- Dumasia, K.; Kumar, A.; Deshpande, S.; Sonawane, S.; Balasinor, N.H. Differential roles of estrogen receptors, ESR1 and ESR2, in adult rat spermatogenesis. Mol. Cell. Endocrinol. 2016, 428, 89–100. [Google Scholar] [CrossRef]
- Cavalcanti, F.N.; Lucas, T.; Lazari, M.; Porto, C.S. Estrogen receptor ESR1 mediates activation of ERK1/2, CREB, and ELK1 in the corpus of the epididymis. J. Mol. Endocrinol. 2015, 54, 339–349. [Google Scholar] [CrossRef]
- Joseph, A.; Hess, R.A.; Schaeffer, D.J.; Ko, C.; Hudgin-Spivey, S.; Chambon, P.; Shur, B.D. Absence of Estrogen Receptor Alpha Leads to Physiological Alterations in the Mouse Epididymis and Consequent Defects in Sperm Function. Biol. Reprod. 2010, 82, 948–957. [Google Scholar] [CrossRef]
- Hess, R.A.; Cooke, P.S. History of estrogen in the male: An historical perspective. Biol. Reprod. 2018, 99, 27–44. [Google Scholar] [CrossRef]
| Names | Primer Sequences (5’→3’) | Product Sizes (bp) | Annealing (Tm, °C) | Notes |
|---|---|---|---|---|
| T1 | TTGCATAGGCAAGCTTTTGG | 1216 | 60 | Clone |
| ATTTTTATCCCCAGACCCTCC |
| Element | Concentration | Direction (µL) |
|---|---|---|
| 2 × Taq Mix | 2× | 12.5 |
| Forward Primer | 100 pmol/L | 1 |
| Reverse Primer | 100 pmol/L | 1 |
| cDNA | 500 ng | 1 |
| ddH2O | 9.5 | |
| Total | 25 |
| Project | Temperature/°C | Time | Cycles |
|---|---|---|---|
| Pre-denaturation | 94 | 2 | ×1 cycles |
| Denaturation | 94 | 1 | ×35 cycles |
| Annealing | 60 | 1 | |
| Extension | 72 | 2 | |
| Extension | 72 | 5 | ×1 cycles |
| Save | 4 | ∞ | ×1 cycles |
| Group | MT950343 | MT950338 | MT950339 | MT950340 | MT950341 | MT950342 |
|---|---|---|---|---|---|---|
| 6 m | 0 | 0 | 0 | 0 | 0 | 50 |
| 18 m | 0 | 0 | 0 | 0 | 2 | 48 |
| 30 m | 33 | 2 | 12 | 3 | 0 | 0 |
| 5 Y | 33 | 1 | 12 | 4 | 0 | 0 |
| Sample | The Score Figure Number | Identify the Number of Spectrograms | Spectral Resolution (%) | Identify the Number of Peptides * | Identifying Protein Number | Unique-2 ** |
|---|---|---|---|---|---|---|
| GST | 4921 | 720 | 14.63 | 180 | 23 | 21 |
| GST_TSSK4 | 11459 | 3828 | 33.41 | 1285 | 241 | 168 |
| Protein ID | Coverage (%) | Mass (Da) | Unique Peptide |
|---|---|---|---|
| tr|A0A6B0QPS2|A0A6B0QPS2_9CETA | 76.44 | 26,849.8 | 27 |
| tr|L8HWB9|L8HWB9_9CETA | 61.34 | 22,418.0 | 12 |
| tr|A0A6B0SBF2|A0A6B0SBF2_9CETA | 48.80 | 41,736.4 | 6 |
| tr|L8I5A7|L8I5A7_9CETA | 51.43 | 23,580.9 | 14 |
| tr|A0A6B0QRL7|A0A6B0QRL7_9CETA | 62.39 | 25,634.6 | 8 |
| GST-TSSK4 | 10.93 | 65,318.1 | 10 |
| tr|L8HP74|L8HP74_9CETA | 17.82 | 51,866.2 | 6 |
| tr|L8IU57|L8IU57_9CETA | 45.05 | 26,112.9 | 8 |
| tr|L8HXW6|L8HXW6_9CETA | 8.01 | 66,485.0 | 2 |
| tr|A0A6B0S215|A0A6B0S215_9CETA | 2.56 | 240,437.2 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Pei, J.; Xiong, L.; Guo, S.; Cao, M.; Kang, Y.; Bao, P.; Wu, X.; Chu, M.; Liang, C.; et al. Identification of the TSSK4 Alternative Spliceosomes and Analysis of the Function of the TSSK4 Protein in Yak (Bos grunniens). Animals 2022, 12, 1380. https://doi.org/10.3390/ani12111380
Wang X, Pei J, Xiong L, Guo S, Cao M, Kang Y, Bao P, Wu X, Chu M, Liang C, et al. Identification of the TSSK4 Alternative Spliceosomes and Analysis of the Function of the TSSK4 Protein in Yak (Bos grunniens). Animals. 2022; 12(11):1380. https://doi.org/10.3390/ani12111380
Chicago/Turabian StyleWang, Xingdong, Jie Pei, Lin Xiong, Shaoke Guo, Mengli Cao, Yandong Kang, Pengjia Bao, Xiaoyun Wu, Min Chu, Chunnian Liang, and et al. 2022. "Identification of the TSSK4 Alternative Spliceosomes and Analysis of the Function of the TSSK4 Protein in Yak (Bos grunniens)" Animals 12, no. 11: 1380. https://doi.org/10.3390/ani12111380
APA StyleWang, X., Pei, J., Xiong, L., Guo, S., Cao, M., Kang, Y., Bao, P., Wu, X., Chu, M., Liang, C., Yan, P., & Guo, X. (2022). Identification of the TSSK4 Alternative Spliceosomes and Analysis of the Function of the TSSK4 Protein in Yak (Bos grunniens). Animals, 12(11), 1380. https://doi.org/10.3390/ani12111380






