Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. siRNAs and Vectors
2.3. Cell Culture and Treatment
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Western Blot
2.6. Dual-Luciferase Assay
2.7. 5-Ethynyl-2′-Deoxyuridine (EdU) Assays
2.8. Flow Cytometry Analysis
2.9. Immunofluorescence
2.10. Statistical Analysis
3. Results
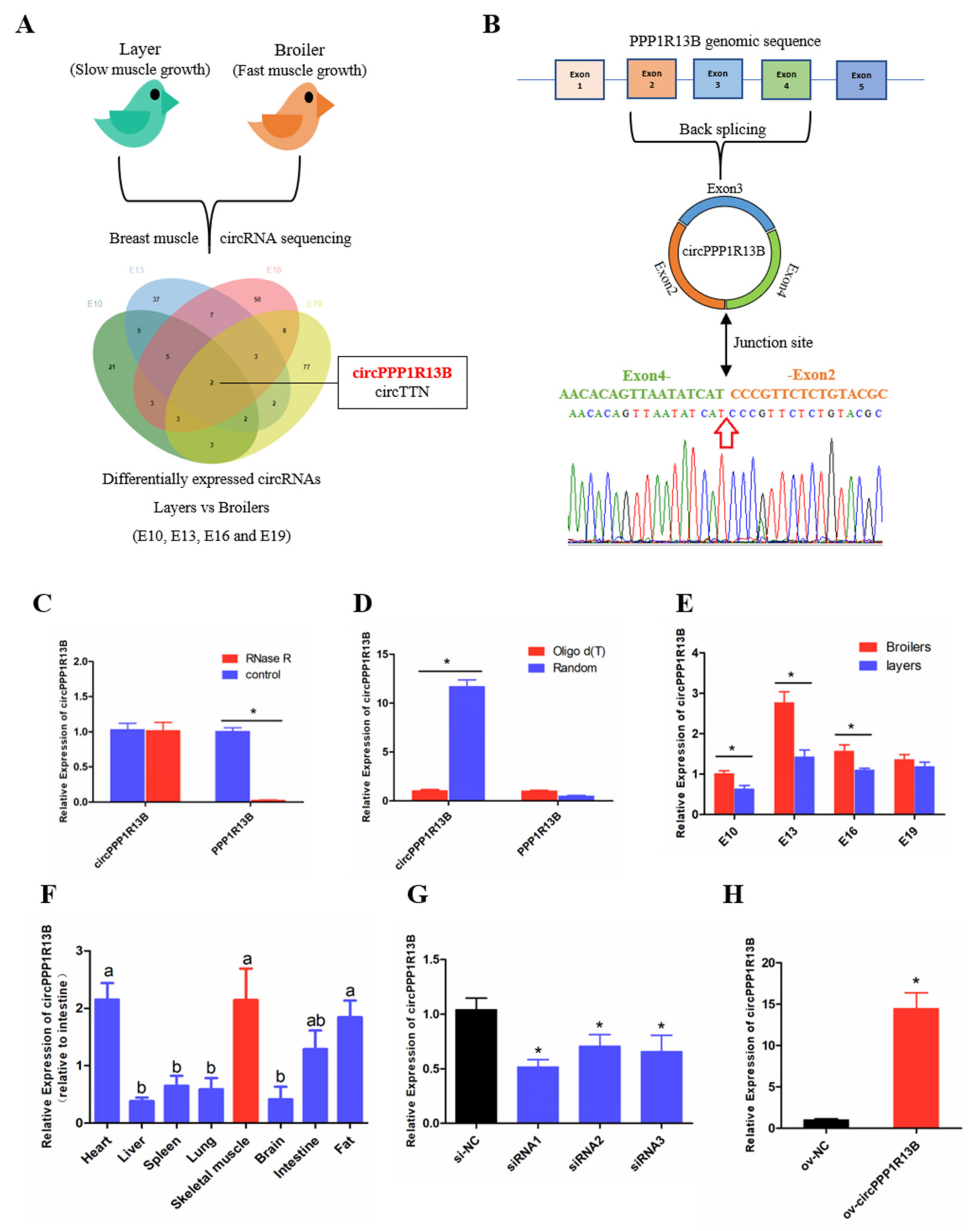
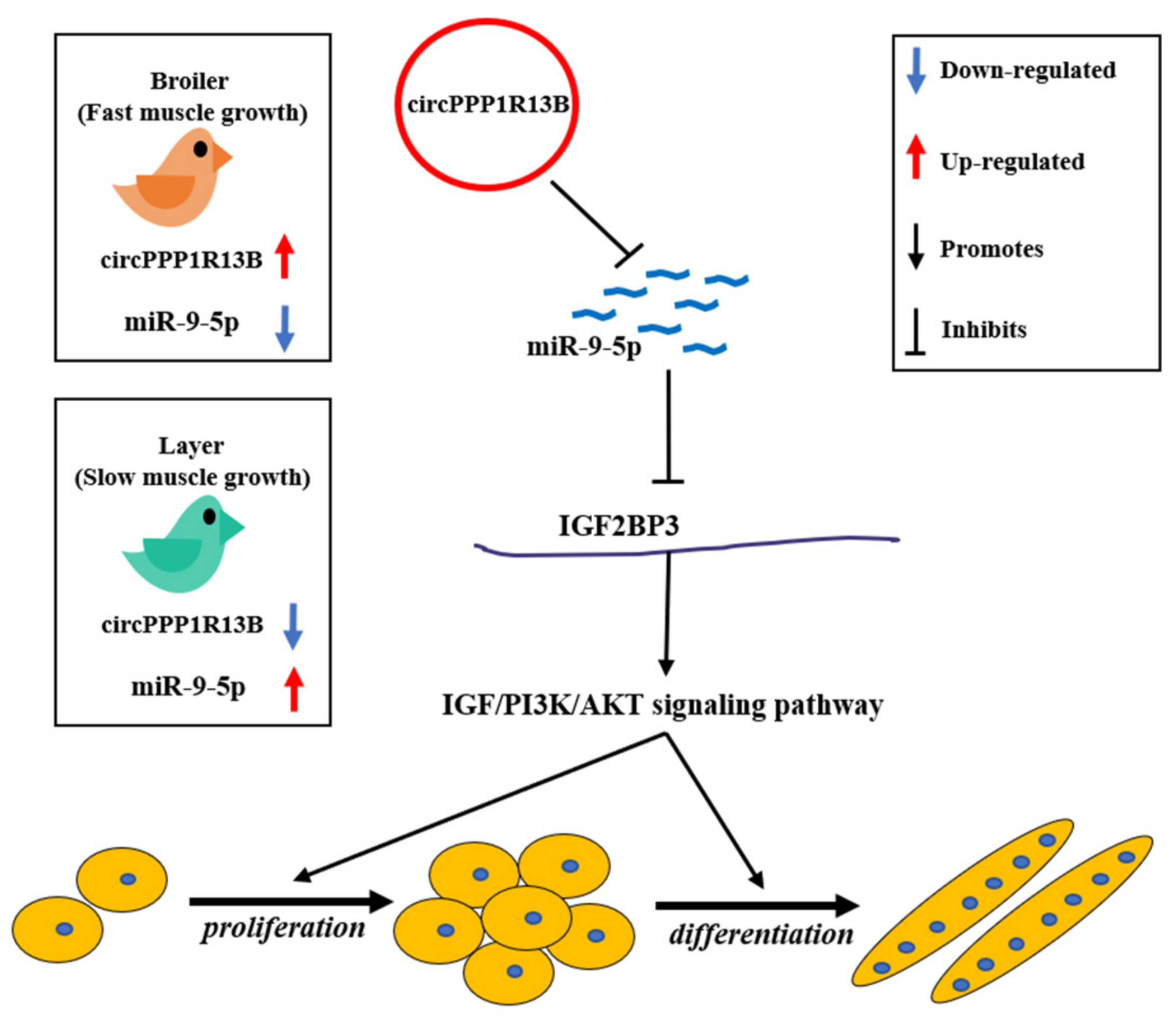
3.1. Circular PPP1R13B RNA Is Differentially Expressed between FMGB and SMGL
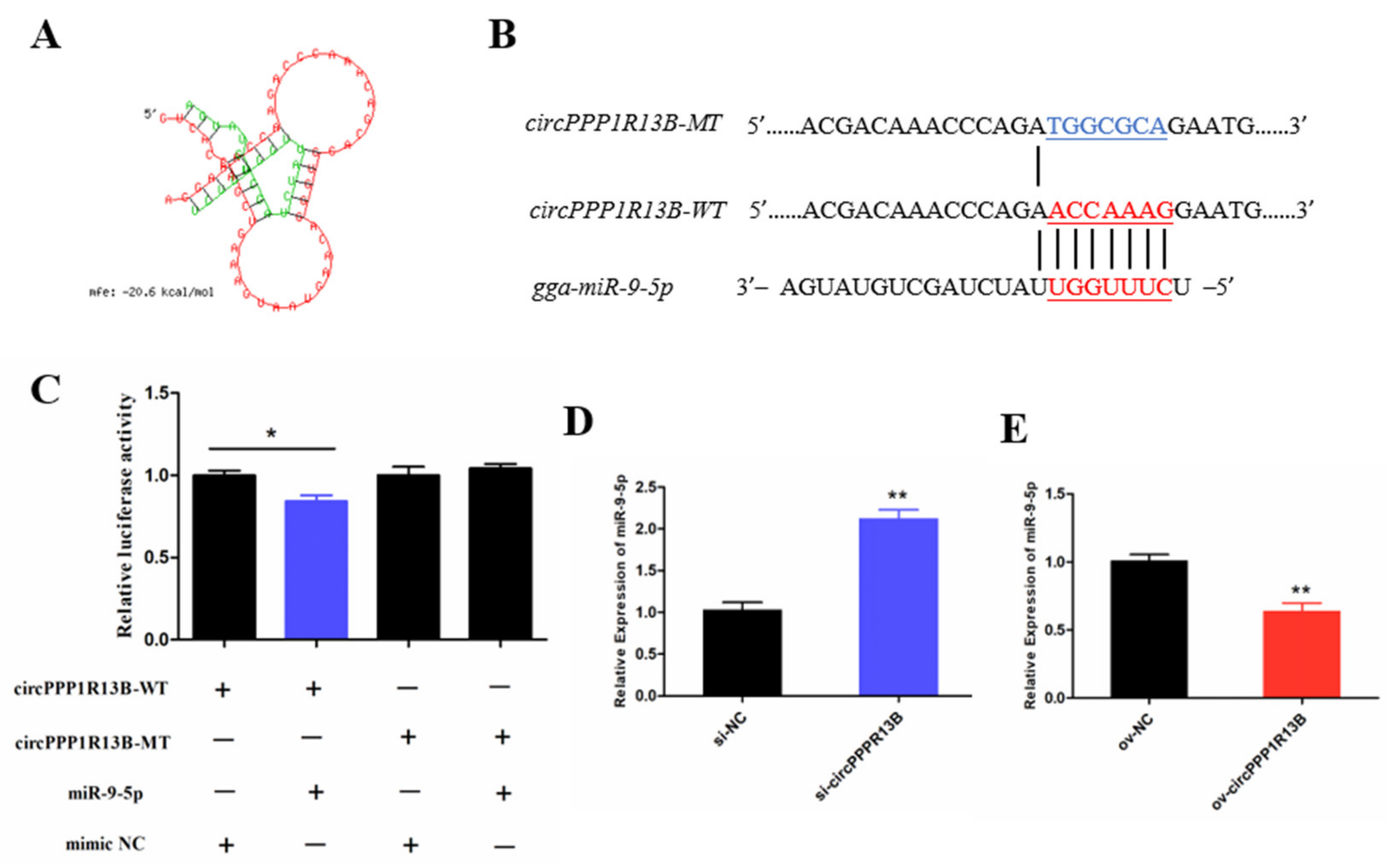
3.2. CircPPP1R13B Interacts with miR-9-5p in Chicken SMSCs
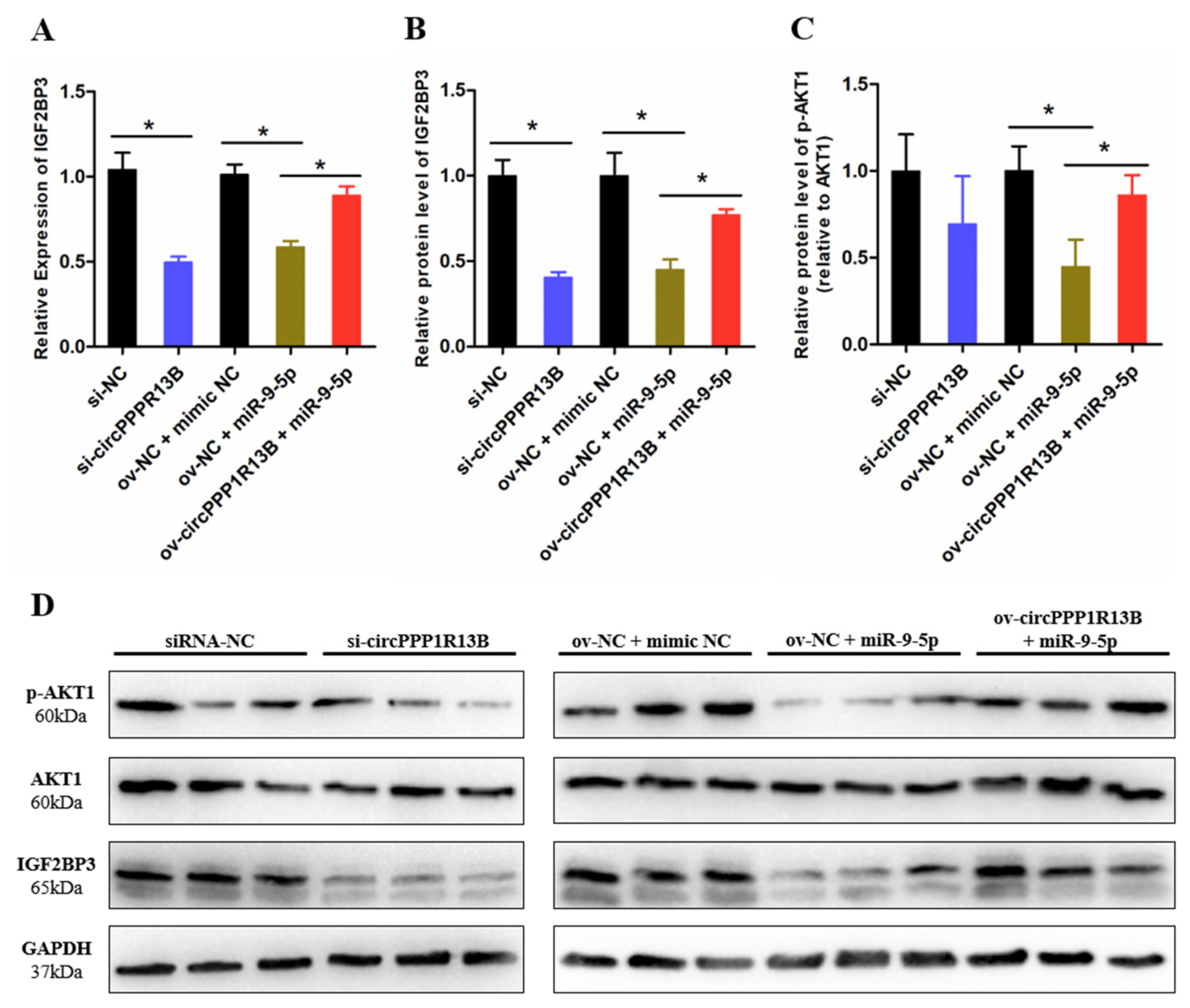
3.3. CircPPP1R13B Regulates IGF2BP3 via Targeting miR-9-5p in Chicken SMSCs
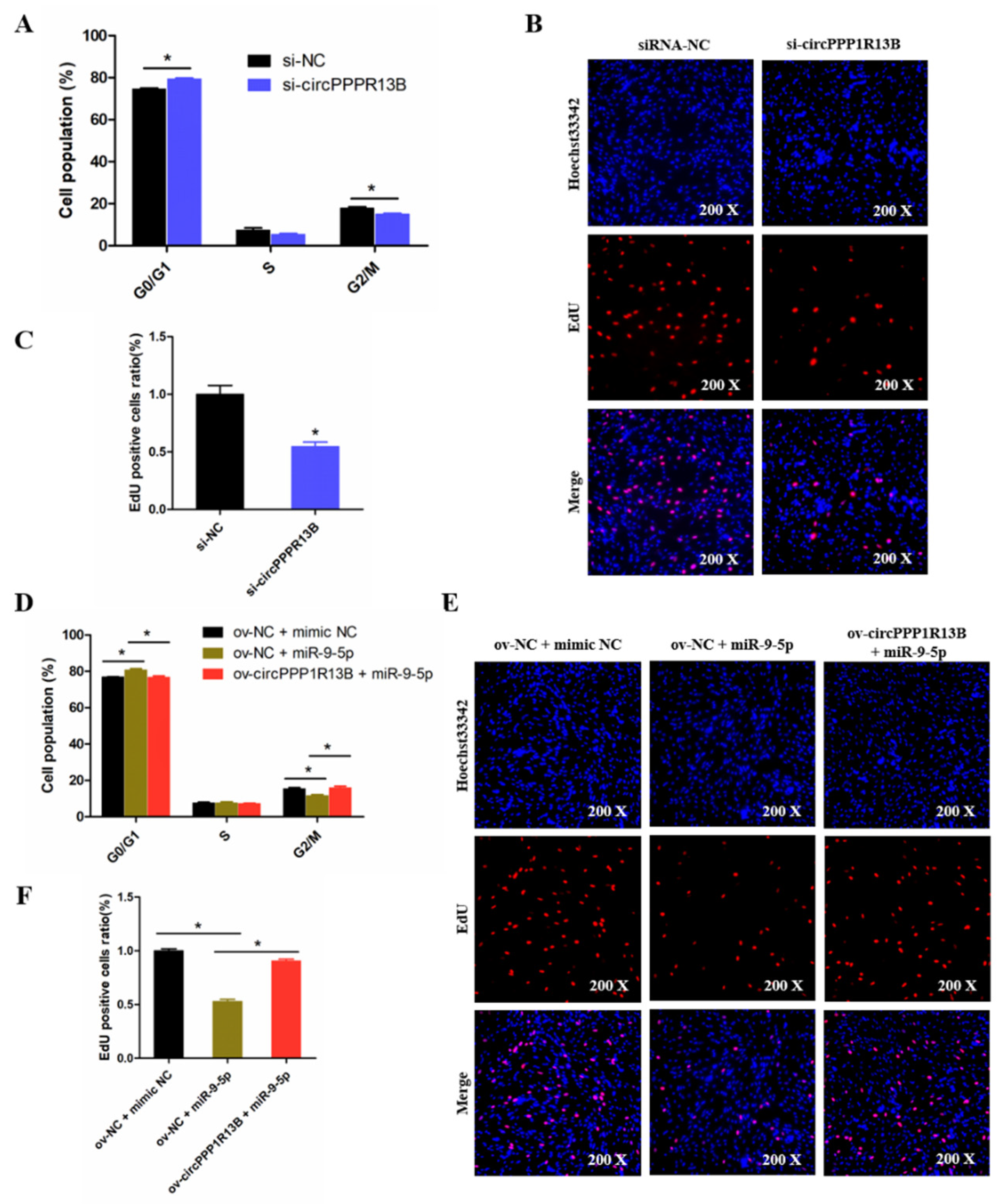
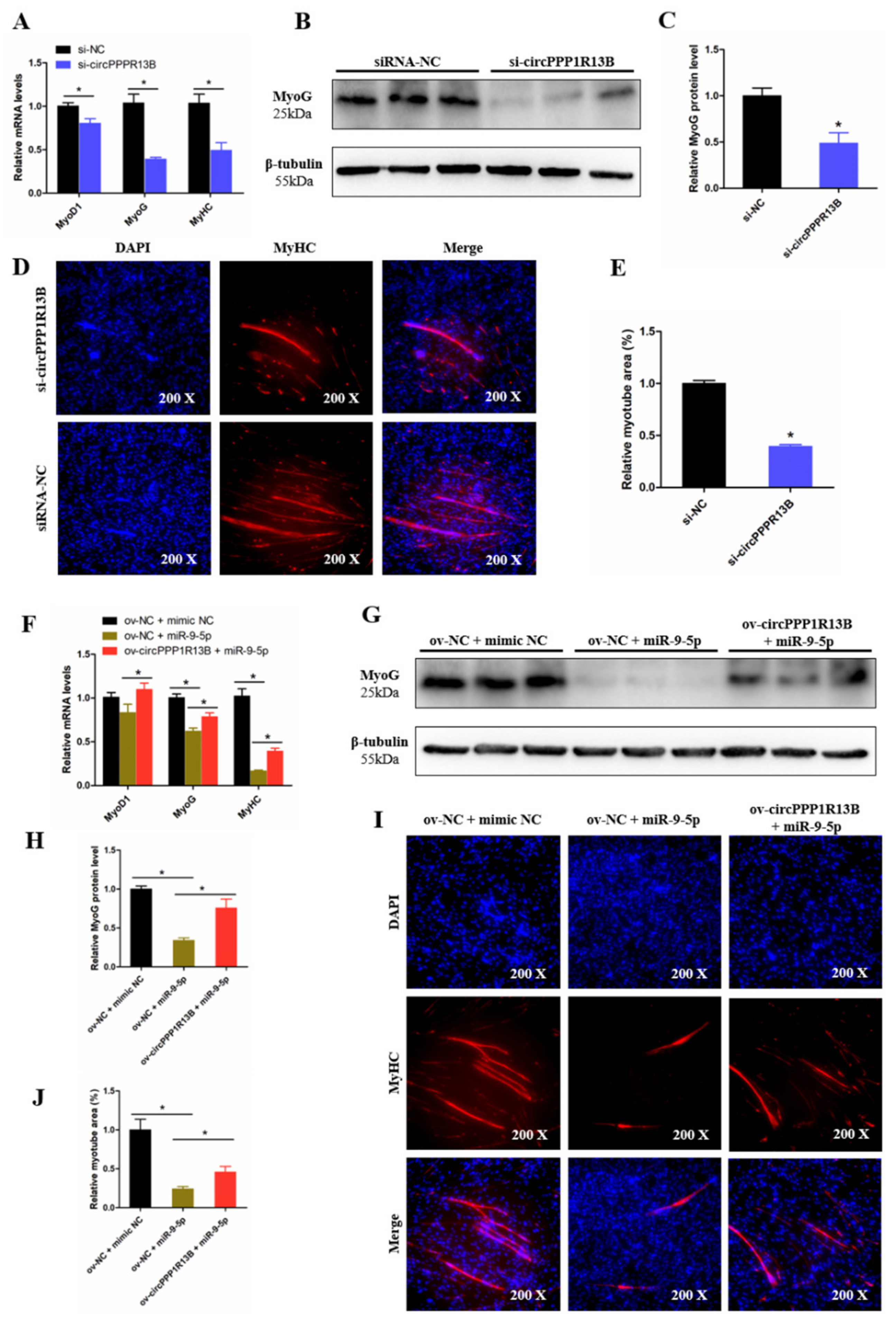
3.4. CircPPP1R13B Eliminates the Inhibition Effect of miR-9-5p on Chicken SMSC Proliferation
3.5. CircPPP1R13B Attenuates the Inhibition Effect of miR-9-5p on Chicken SMSC Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Du, Y.; Liu, L.; He, Y.; Dou, T.; Jia, J.; Ge, C. Endocrine and genetic factors affecting egg laying performance in chickens: A review. Br. Poult. Sci. 2020, 61, 538–549. [Google Scholar] [CrossRef]
- Aiello, D.; Patel, K.; Lasagna, E. The myostatin gene: An overview of mechanisms of action and its relevance to livestock animals. Anim. Genet. 2018, 49, 505–519. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, R.; Wei, Y.; Meng, X.; Wang, B.; Zhang, Z.; Wu, W.; Liu, H. Effect of mstn mutation on growth and carcass performance in duroc x meishan hybrid population. Animals 2020, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Li, C. Growth hormone receptor mutations related to individual dwarfism. Int. J. Mol. Sci. 2018, 19, 1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.X.; Shen, Q.C.; Zheng, M.Q.; Su, Y.C.; Cai, R.C.; Yu, Y.; Yang, X.R.; Chen, Z.W.; Wen, J.; Zhao, G.P. A selection method of chickens with blue-eggshell and dwarf traits by molecular marker-assisted selection. Poult. Sci. 2019, 98, 3114–3118. [Google Scholar] [CrossRef] [PubMed]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key genes regulating skeletal muscle development and growth in farm animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Le Grand, F.; Rudnicki, M.A. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 2007, 19, 628–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.V.; Hughes, S.M. Mef2 and the Skeletal Muscle Differentiation Program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Zanou, N.; Gailly, P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (mrfs) and insulin-like growth factors (igfs) pathways. Cell Mol. Life Sci. CMLS 2013, 70, 4117–4130. [Google Scholar] [CrossRef]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circ rna s. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular rna molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Ji, P.; Zhao, F. Circatlas: An integrated resource of one million highly accurate circular rnas from 1070 vertebrate transcriptomes. Genome Biol. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.-L. The biogenesis, functions, and challenges of circular rnas. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, Y.; Nie, L.; Ding, X.; Zhang, X.; Zhao, W.; Xu, X.; Kyei, B.; Dai, D.; Zhan, S. Myod-induced circular rna cdr1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 807–821. [Google Scholar] [CrossRef]
- Gao, M.; Li, X.; Yang, Z.; Zhao, S.; Ling, X.; Li, J.; Xing, K.; Qi, X.; Wang, X. Circhipk3 regulates proliferation and differentiation of myoblast through the mir-7/tcf12 pathway. J. Cell. Physiol. 2021. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Kong, L.; Cao, M.; Zhang, M.; Wang, Y.; Song, C.; Fang, X.; Chen, H.; Zhang, C. Circarid1a regulates mouse skeletal muscle regeneration by functioning as a sponge of mir-6368. FASEB J. 2021, 35, e21324. [Google Scholar]
- Shen, X.; Tang, J.; Jiang, R.; Wang, X.; Yang, Z.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Circrilpl1 promotes muscle proliferation and differentiation via binding mir-145 to activate igf1r/pi3k/akt pathway. Cell Death Dis. 2021, 12, 142. [Google Scholar] [CrossRef]
- Griffin, H.D.; Goddard, C. Rapidly growing broiler (meat-type) chickens: Their origin and use for comparative studies of the regulation of growth. Int. J. Biochem. 1994, 26, 19–28. [Google Scholar] [CrossRef]
- Aberle, E.; Stewart, T. Growth of fiber types and apparent fiber number in skeletal muscle of broiler-and layer-type chickens. Growth 1983, 47, 135–144. [Google Scholar]
- Shen, X.; Liu, Z.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Zhao, J.; Li, D.; Wang, Y. Circular rna profiling identified an abundant circular rna circtmtc1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging mir-128-3p. Int. J. Biol. Sci. 2019, 15, 2265. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; He, H.; Cao, X.; Shen, X.; Han, S.; Cui, C.; Zhao, J.; Wei, Y.; Chen, Y.; Xia, L. Mir-148a-3p regulates skeletal muscle satellite cell differentiation via the pi3k/akt signaling pathway by targeting meox2. Front. Genet. 2020, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shen, X.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Wei, Y.; Wang, Y.; Li, D. Hdac4 regulates the proliferation, differentiation and apoptosis of chicken skeletal muscle satellite cells. Animals 2020, 10, 84. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular rnas are a large class of animal rnas with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Yin, H.; He, H.; Shen, X.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y.; Xia, L. Mir-9-5p inhibits skeletal muscle satellite cell proliferation and differentiation by targeting igf2bp3 through the igf2-pi3k/akt signaling pathway. Int. J. Mol. Sci. 2020, 21, 1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, H.; He, X.; Li, G.; Xu, H.; Jia, X.; Nie, Q.; Zhang, X. Deep sequencing analysis of mirna expression in breast muscle of fast-growing and slow-growing broilers. Int. J. Mol. Sci. 2015, 16, 16242–16262. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; He, H.; Shen, X.; Tang, S.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y. Microrna profiling reveals an abundant mir-200a-3p promotes skeletal muscle satellite cell development by targeting tgf-β2 and regulating the tgf-β2/smad signaling pathway. Int. J. Mol. Sci. 2020, 21, 3274. [Google Scholar] [CrossRef]
- Das, A.; Das, A.; Das, D.; Abdelmohsen, K.; Panda, A.C. Circular rnas in myogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194372. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Chen, X.; Li, W.; Li, Z.; Nie, Q.; Zhang, X. Circular rna circsvil promotes myoblast proliferation and differentiation by sponging mir-203 in chicken. Front. Genet. 2018, 9, 172. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ouyang, H.; Wang, Z.; Chen, B.; Nie, Q. A novel circular rna generated by fgfr2 gene promotes myoblast proliferation and differentiation by sponging mir-133a-5p and mir-29b-1-5p. Cells 2018, 7, 199. [Google Scholar] [CrossRef] [Green Version]
- Yue, B.; Wang, J.; Song, C.; Wu, J.; Cao, X.; Huang, Y.; Lan, X.; Lei, C.; Huang, B.; Chen, H. Biogenesis and cerna role of circular rnas in skeletal muscle myogenesis. Int. J. Biochem. Cell Biol. 2019, 117, 105621. [Google Scholar] [CrossRef]
- Lin, S.; Luo, W.; Ye, Y.; Bekele, E.J.; Nie, Q.; Li, Y.; Zhang, X. Let-7b regulates myoblast proliferation by inhibiting igf2bp3 expression in dwarf and normal chicken. Front. Physiol. 2017, 8, 477. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Delafontaine, P. Mechanisms of igf-1-mediated regulation of skeletal muscle hypertrophy and atrophy. Cells 2020, 9, 1970. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cai, B.; Abdalla, B.A.; Zhu, X.; Zheng, M.; Han, P.; Nie, Q.; Zhang, X. Lncirs1 controls muscle atrophy via sponging mir-15 family to activate igf1-pi3k/akt pathway. J. Cachexia Sarcopenia Muscle 2019, 10, 391–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Cao, X.; Dong, D.; Shen, X.; Cheng, J.; Jiang, R.; Yang, Z.; Peng, S.; Huang, Y.; Lan, X. Circular rna ttn acts as a mir-432 sponge to facilitate proliferation and differentiation of myoblasts via the igf2/pi3k/akt signaling pathway. Mol. Ther. Nucleic Acids 2019, 18, 966–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Wei, Y.; You, G.; Liu, W.; Amevor, F.K.; Zhang, Y.; He, H.; Ma, M.; Zhang, Y.; Li, D.; et al. Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p. Animals 2021, 11, 2396. https://doi.org/10.3390/ani11082396
Shen X, Wei Y, You G, Liu W, Amevor FK, Zhang Y, He H, Ma M, Zhang Y, Li D, et al. Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p. Animals. 2021; 11(8):2396. https://doi.org/10.3390/ani11082396
Chicago/Turabian StyleShen, Xiaoxu, Yuanhang Wei, Guishuang You, Wei Liu, Felix Kwame Amevor, Yao Zhang, Haorong He, Menggen Ma, Yun Zhang, Diyan Li, and et al. 2021. "Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p" Animals 11, no. 8: 2396. https://doi.org/10.3390/ani11082396
APA StyleShen, X., Wei, Y., You, G., Liu, W., Amevor, F. K., Zhang, Y., He, H., Ma, M., Zhang, Y., Li, D., Zhu, Q., & Yin, H. (2021). Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p. Animals, 11(8), 2396. https://doi.org/10.3390/ani11082396






