Comparative Expression Profiling and Sequence Characterization of ATP1A1 Gene Associated with Heat Tolerance in Tropically Adapted Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Quantitative Real Time PCR (q-RT PCR)
2.2. Amplification of the ATP1A1 Gene
2.3. Sequence, Molecular Clock, and Data Analysis
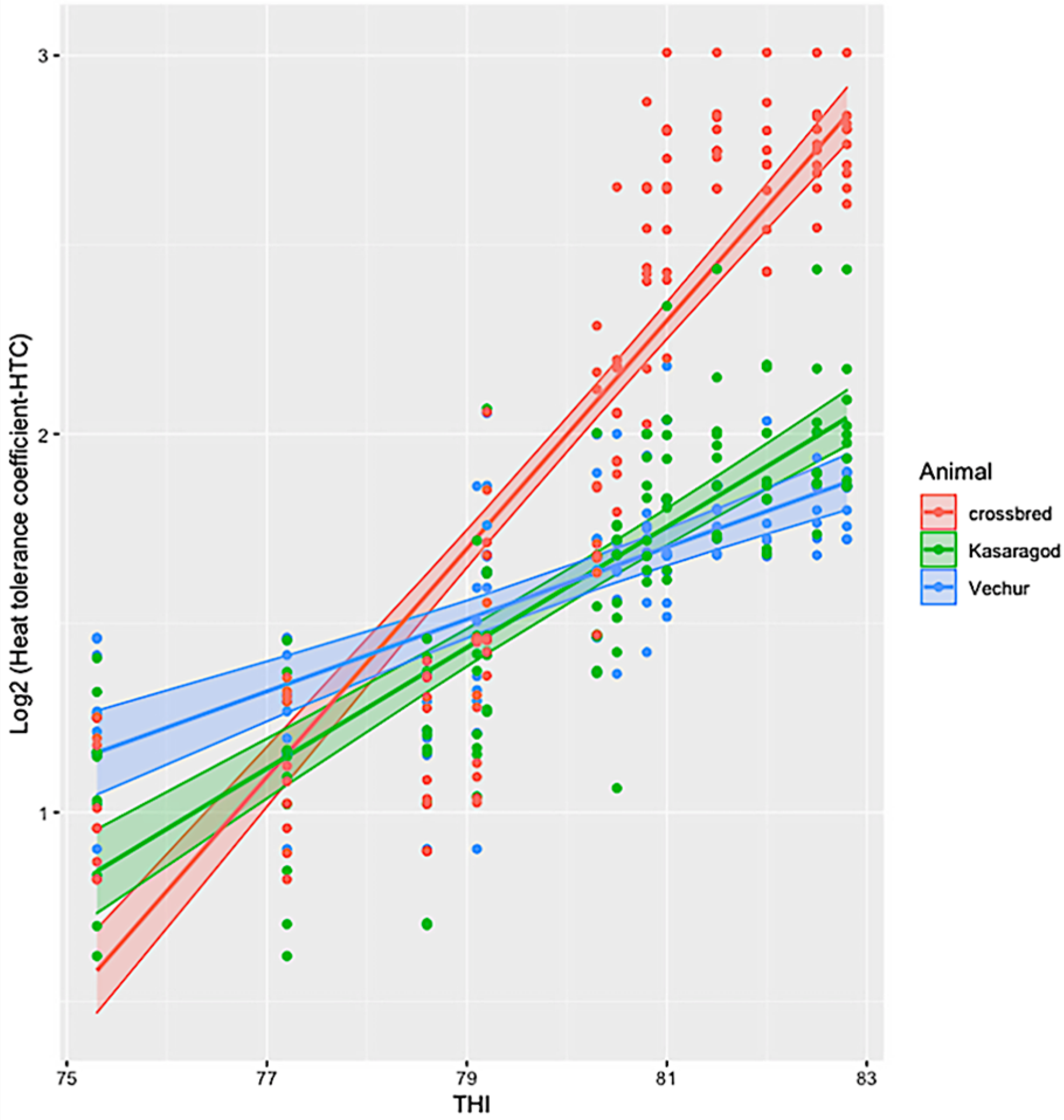
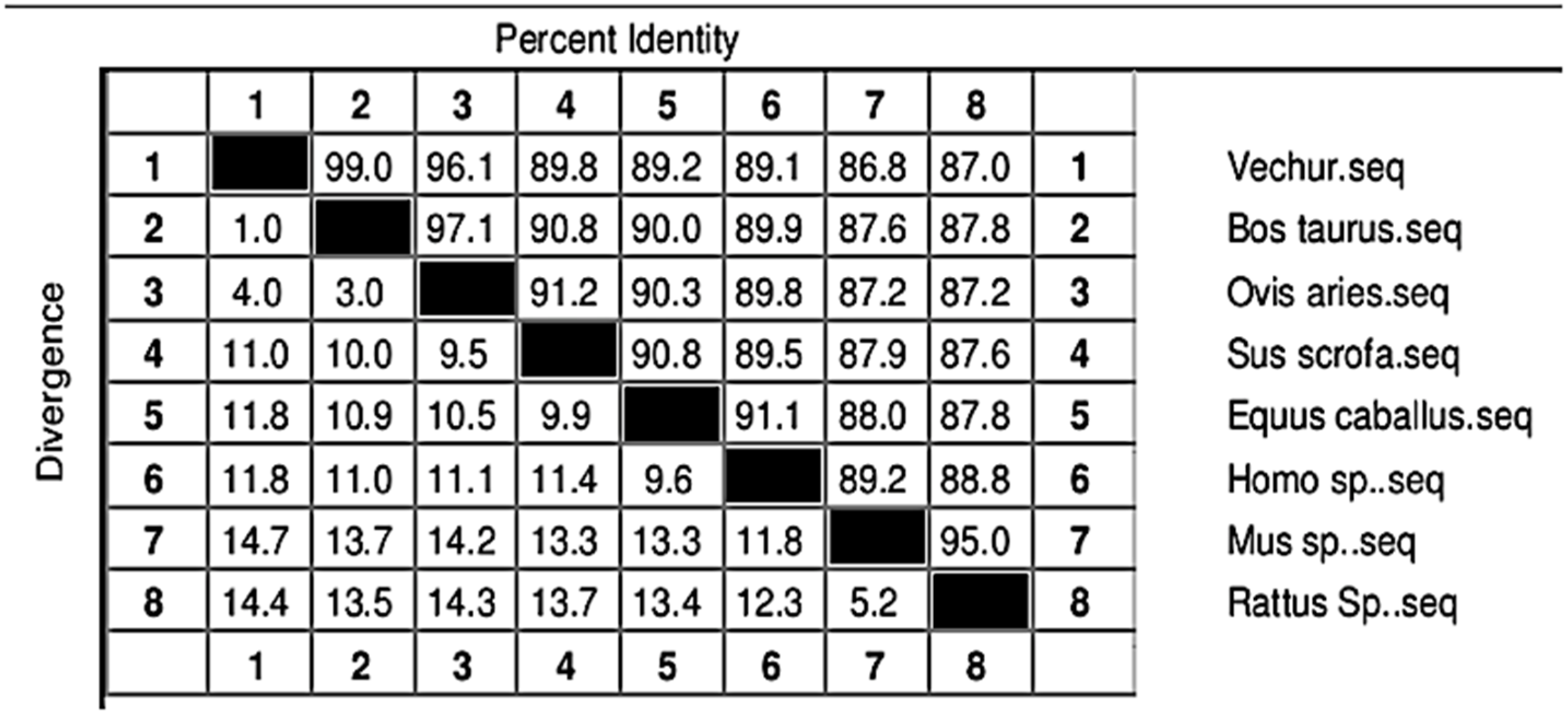
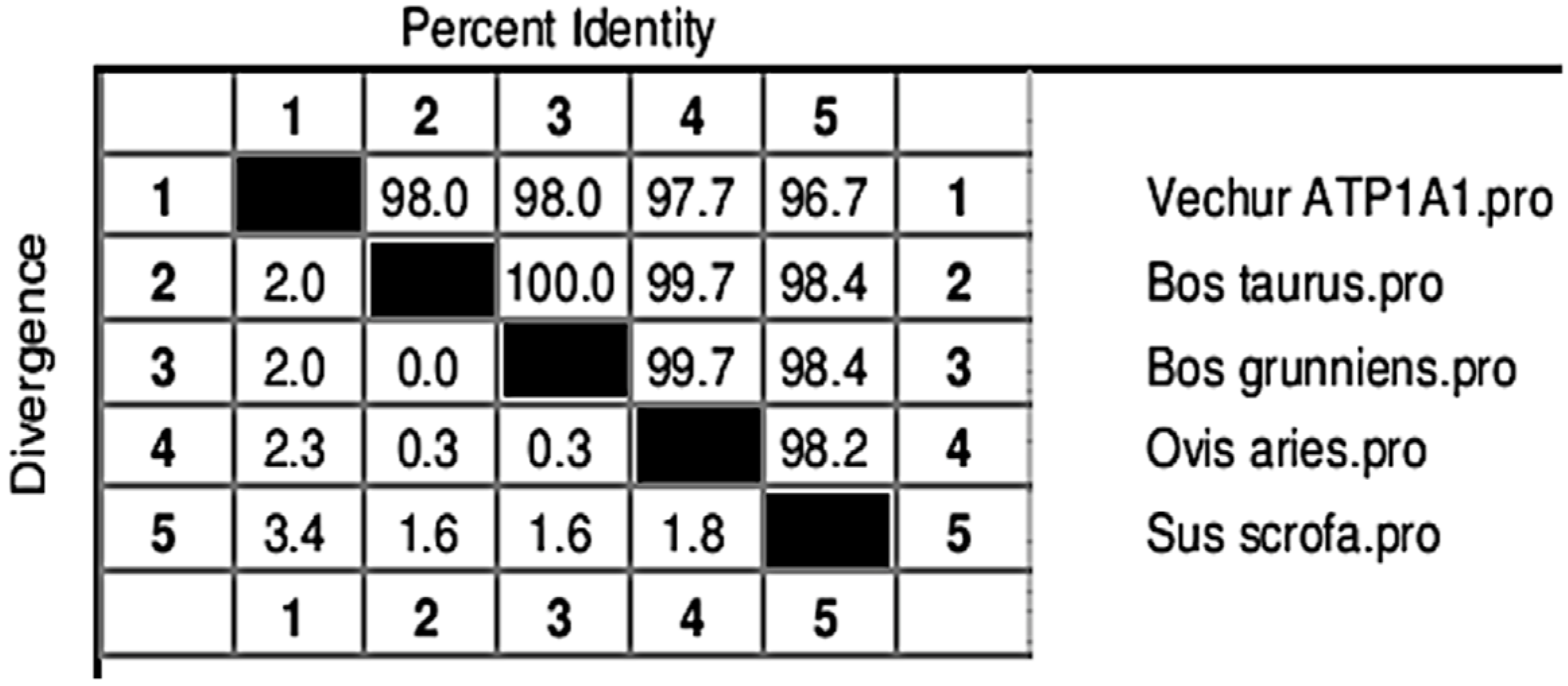
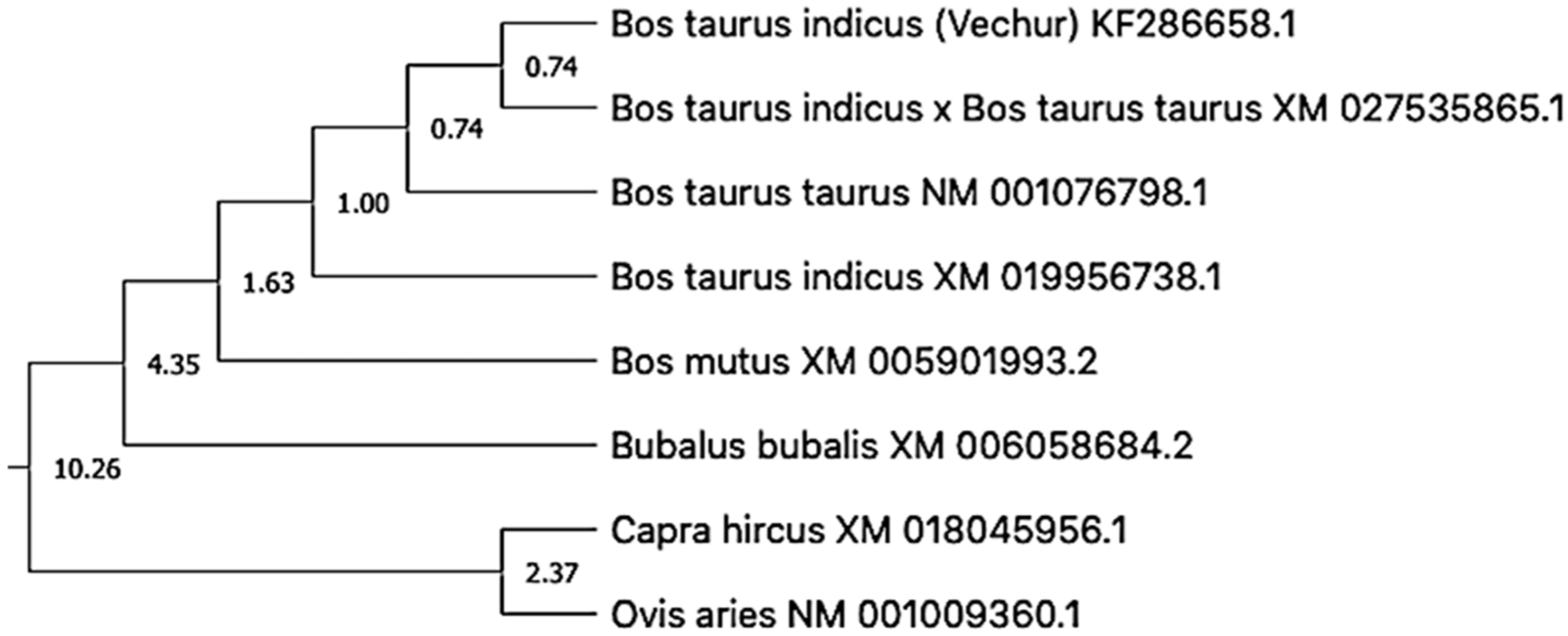
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eisler, M.C.; Lee, M.R.F.; Tarlton, J.F.; Martin, G.B.; Beddington, J.; Dungait, J.A.; Greathead, H.; Liu, J.; Mathew, S.; Miller, H.; et al. Agriculture: Steps to sustainable livestock. Nature 2014, 507, 32–34. [Google Scholar] [CrossRef]
- Van Zanten, H.H.; Herrero, M.; Van Hal, O.; Roos, E.; Muller, A.; Garnett, T.; Gerber, P.J.; Schader, C.; De Boer, I.J. Defining a land boundary for sustainable livestock consumption. Glob. Chang. Biol. 2018, 24, 4185–4194. [Google Scholar] [CrossRef] [PubMed]
- Bromham, L.; Penny, D. The modern molecular clock. Nat. Rev. Genet. 2003, 4, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Huang, B.; Zhan, J.; Wang, J.; Qu, K.; Zhang, F.; Shen, J.; Jia, P.; Ning, Q.; Zhang, J.; et al. Whole-genome analyses identify loci and selective signals associated with body size in cattle. J. Anim. Sci. 2020, 98, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Perini, F.; Cendron, F.; Rovelli, G.; Castellini, C.; Cassandro, M.; Lasagna, E. Emerging genetic tools to investigate molecular pathways related to heat stress in chickens: A review. Animals 2021, 11, 1–19. [Google Scholar]
- Elayadeth-Meethal, M.; ThazhathuVeettil, A.; Maloney, S.K.; Hawkins, N.; Misselbrook, T.H.; Sejian, V.; Rivero, M.J.; Lee, M.R. Size does matter: Parallel evolution of adaptive thermal tolerance and body size facilitates adaptation to climate change in domestic cattle. Ecol. Evol. 2018, 8, 10608–10620. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.Z.; Sasazaki, S.; Mannen, H. Genetic diversity and structure in Bos taurus and Bos indicus populations analyzed by SNP markers. Anim. Sci. 2010, 81, 281–289. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Kemper, K.E.; Hayes, B.J.; Zhang, Y.; Tier, B.; Barendse, W.; Reverter, A.; Goddard, M.E. Detection of quantitative trait loci in Bos indicus and Bos taurus cattle using genome-wide association studies. Genet. Sel. Evol. 2013, 45, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; Stiening, C.M.; Pollard, B.C.; Vanbaale, M.J.; Baumgard, L.H.; Gentry, P.C.; Coussens, P.M. Use of expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J. Anim. Sci. 2006, 84, 1–13. [Google Scholar] [CrossRef]
- Collier, R.J.; Collier, J.L.; Rhoads, R.P.; Baumgard, L.H. Genes involved in the bovine heat stress response. J. Dairy Sci. 2008, 91, 445–454. [Google Scholar] [CrossRef]
- Agnew, L.; Colditz, I.G. Development of a method of measuring cellular stress in cattle and sheep. Vet. Immunl. Immunopathol. 2008, 123, 197–204. [Google Scholar] [CrossRef]
- Rajamanickam, G.D.; Kastelic, J.P.; Thundathil, J.C. Na/K-ATPase regulates bovine sperm capacitation through raft-and non-raft-mediated signaling mechanisms. Mol. Reprod. Dev. 2017, 84, 1168–1182. [Google Scholar] [CrossRef]
- Mavrogonatou, E.; Papadimitriou, K.; Urban, J.P.; Papadopoulos, V.; Kletsas, D. Deficiency in the α1 subunit of Na+/K+-ATPase enhances the anti-proliferative effect of high osmolality in nucleus pulposus intervertebral disc cells. J. Cell. Physiol. 2015, 230, 3037–3048. [Google Scholar] [CrossRef]
- Liu, Y.X.; Xu, C.H.; Gao, T.Y.; Sun, Y. Polymorphisms of the ATP1A1 gene associated with mastitis in dairy cattle. Genet. Mol. Res. 2012, 11, 651–660. [Google Scholar] [CrossRef]
- Geering, K.; Kraehenbuhl, J.P.; Rossier, B.C. Maturation of the catalytic alpha-subunit of Na, K- ATPase during intracellular transport. J. Cell Biol. 1987, 105, 2613–2619. [Google Scholar] [CrossRef]
- Das, R.; Gupta, I.D.; Verma, A.; Singh, S.; Chaudhari, M.V.; Sailo, L.; Verma, N.; Kumar, R. Single nucleotide polymorphisms in ATP1A1gene and their association with thermotolerance traits in Sahiwal and Karan Fries cattle. India. J. Anim. Res. 2017, 51, 70–74. [Google Scholar]
- Wang, Z.; Huang, J.; Zhong, J.; Wang, G. Tissue specific alternative splicing and expression of ATP1B2 gene. Afr. J. Biotech. 2012, 39, 9485–9495. [Google Scholar]
- Wang, S.; Edens, F.W. Involvement of steroid hormones, corticosterone and testosterone, in synthesis of heat shock proteins in broiler chicken. Int. J. Poult. Sci. 2008, 7, 783–797. [Google Scholar] [CrossRef]
- Wang, L.; Schumann, U.; Liu, Y.; Prokopchuk, O.; Steinacker, J.M. Heat shock protein 70 (Hsp70) inhibits oxidative phosphorylation and compensates ATP balance through enhanced glycolytic activity. J. Appl. Physiol. 2012, 113, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S.; et al. Whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Zhou, X.; Li, D.Q.; Cui, Q.W.; Wang, G.L. Association of ATP 1 A 1 gene polymorphism with heat tolerance traits in dairy cattle. Genet. Mol. Res. 2010, 9, 891–896. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Li, H.; Zhou, X.; Wang, G. A Novel SNP of the ATP1A1 gene is associated with heat tolerance traits in dairy cows. Mol. Biol. Rep. 2011, 38, 83–88. [Google Scholar] [CrossRef]
- Vague, P.; Dufayet, D.; Coste, T.; Moriscot, C.; Jannot, M.F.; Raccah, D. Association of diabetic neuropathy with Na/K ATPase gene polymorphism. Diabetologia 1997, 40, 506–511. [Google Scholar] [CrossRef]
- Schlingmann, K.P.; Bandulik, S.; Mammen, C.; Tarailo-Graovac, M.; Holm, R.; Baumann, M.; Konig, J.; Lee, J.J.; Drogemoller, B.; Imminger, K.; et al. Germline de novo mutations in ATP1A1 cause renal hypomagnesemia, refractory seizures, and intellectual disability. Am. J. Hum. Genet. 2018, 103, 808–816. [Google Scholar] [CrossRef]
- Stregapede, F.; Travaglini, L.; Rebelo, A.P.; Cintra, V.P.; Bellacchio, E.; Bosco, L.; Alfieri, P.; Pro, S.; Zuchner, S.; Bertini, E.; et al. Hereditary spastic paraplegia is a novel phenotype for germline de novo ATP1A1 mutation. Clin. Genet. 2020, 97, 521–526. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.P.; Sanchez, G.; Mitra, A.; Numata, S.; Liu, L.C.; Blanco, G. Na, K-ATPase α4, and not Na, K-ATPase α1, is the main contributor to sperm motility, but its high ouabain binding affinity site is not required for male fertility in mice. J. Membr. Biol. 2021, 1–13. [Google Scholar]
- Das, R.; Gupta, I.D.; Verma, A.; Singh, A.; Chaudhari, M.V.; Sailo, L.; Upadhyay, R.C.; Goswami, J. Genetic polymorphisms in ATP1A1 gene and their association with heat tolerance in Jersey crossbred cows. India. J. Dairy Sci. 2015, 68, 50–54. [Google Scholar]
- Ramendra, D.; Gupta, I.D.; Archana, V.; Chaudhari, M.V.; Lalrengpuii, S.; Sohanvir, S. Identification of SNPs in ATP1A1 gene and their association with heat tolerance in Sahiwal and Karan Fries (Bos taurus× Bos indicus) cattle under tropical climatic condition. Indian, J. Dairy Sci. 2018, 71, 409–415. [Google Scholar]
- Imran, S.; Khan, M.S.; Qureshi, Z.I. Genetic characterization of Cholistani breed of cattle for ATP1A1 gene and its association to heat tolerance traits. Pak. J. Agric. Sci. 2021, 58, 229–234. [Google Scholar]
- Faheem, M.S.; Ghanem, N.; Gad, A.; Procházka, R.; Dessouki, S.M. Adaptive and biological responses of buffalo granulosa cells exposed to heat stress under In vitro condition. Animals 2021, 11, 794. [Google Scholar] [CrossRef]
- Kashyap, N.; Kumar, P.; Deshmukh, B.; Bhat, S.; Kumar, A.; Chauhan, A.; Bhushan, B.; Singh, G.; Sharma, D. Association of ATP1A1 gene polymorphism with thermotolerance in Tharparkar and Vrindavani cattle. Vet. World 2015, 8, 892–897. [Google Scholar] [CrossRef][Green Version]
- Chang, J.T.; Lowery, L.A.; Sive, H. Multiple roles for the Na, K-ATPase subunits Atp1a1 and Fxyd1, during brain ventricle development. Dev. Biol. 2012, 368, 312–322. [Google Scholar] [CrossRef]
- Thompson, C.B.; Dorup, I.; Ahn, J.; Leong, P.K.; McDonough, A.A. Glucocorticoids increase sodium pump α2-and β1-subunit abundance and mRNA in rat skeletal muscle. Am. J. Physiol. Cell Physiol. 2001, 280, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Burkard, C.; Verheije, M.H.; Haagmans, B.L.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J.; de Haan, C.A. ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J. Virol. 2015, 89, 4434–4448. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.H.; Ellakwa, D.E.S. SUMO pathway, blood coagulation and oxidative stress in SARS-CoV-2 infection. Biochem. Biophys. Rep. 2021, 26, 1–6. [Google Scholar]
- Medina-Ortiz, K.; Lopez-Alvarez, D.; Navia, F.; Hansen, T.; Fierro, L.; Castano, S. Identification of Na+/K+-ATPase α/β isoforms in Rhinella marina tissues by RNAseq and a molecular docking approach at the protein level to evaluate α isoform affinities for bufadienolides. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2021, 254, 1–14. [Google Scholar]
- Yang, N.; Shen, H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef]
- Bohmanova., J.; Misztal, I.; Tsuruta, S.; Norman, H.D.; Lawlor, T.J. Genotype by environment interaction due to heat stress. J. Dairy Sci. 2008, 91, 840–846. [Google Scholar] [CrossRef]
- Tichopad, A.; Kitchen, R.; Riedmaier, I.; Becker, C.; Kubista, M. Design and optimization of reverse transcription quantitative PCR experiments. Clin. Chem. 2009, 55, 1816–1823. [Google Scholar] [CrossRef]
- Xiang-Hong, J.; Hong, Y.Y.; Jin, X.H.; Mei, X.Y.; Rong, J.P.; Ming, L. Selection of reference genes for gene expression studies in PBMC from Bama miniature pig under heat stress. Vet. Immunol. Immunopathol. 2011, 144, 160–166. [Google Scholar] [CrossRef]
- Mahadav, A.; Kontsedelov, S.; Czosnek, H.; Ghanim, M. Thermotolerance and gene expression following heat stress in the whitefly Bamesia tobaci B and Q biotypes. Insect Biochem. Mol. Biol. 2009, 39, 668–676. [Google Scholar] [CrossRef]
- Eitam, H.; Vaya, J.; Brosh, A.; Orlow, A.; Khatib, S.; Izhaki, I.; Shabtay, A. Differential stress responses among newly received calves: Variations in reductant capacity and HSP gene expression. Cell Stress Chaperones 2010, 15, 865–876. [Google Scholar] [CrossRef]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Biol. Direct 2008, 3, 1–13. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef]
- Tamura, K.; Qiqing, T.; Kumar, S. Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Tao, Q.; Tamura, K.; Mello, B.; Kumar, S. Confidence intervals for RelTime estimates of divergence times from molecular data. Mol. Biol. Evol. 2020, 37, 280–290. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Loftus, R.T.; MacHugh, D.E.; Bradley, D.G.; Sharp, P.M.; Cunningham, P. Evidence for two independent domestications of cattle. Proc. Natl. Acad. Sci. USA 1994, 91, 2757–2761. [Google Scholar] [CrossRef]
- MacHugh, D.E.; Shriver, M.D.; Loftus, R.T.; Cunningham, P.; Bradley, D.G. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 1997, 146, 1071–1086. [Google Scholar] [CrossRef]
- Gillooly, J.F.; Allen, A.P.; West, G.B.; Brown, J.H. The rate of DNA evolution: Effects of body size and temperature on the molecular clock. Proc. Natl. Acad. Sci. USA 2005, 102, 140–145. [Google Scholar] [CrossRef]
- Thomas, J.A.; Welch, J.J.; Woolfit, M.; Bromham, L. There is no universal molecular clock for invertebrates, but rate variation does not scale with body size. Proc. Natl. Acad. Sci. USA 2006, 103, 7366–7371. [Google Scholar] [CrossRef] [PubMed]
- Pulquerio, M.J.; Nichols, R.A. Dates from the molecular clock: How wrong can we be? Trends Ecol. Evol. 2007, 22, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.M.; Lajeunesse, M.J.; Rohr, J.R. A global synthesis of animal phenological responses to climate change. Nat. Clim. Chang. 2018, 8, 224–228. [Google Scholar] [CrossRef]
- Vaidya, M.M.; Kumar, P.; Singh, S.V. Circadian changes in heat storage and heat loss through sweating and panting in Karan Fries cattle during different seasons. Biol. Rhythm Res. 2011, 43, 137–146. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I. Genetic component of heat stress in dairy cattle, parameter estimation. J. Dairy Sci. 2000, 83, 2126–2130. [Google Scholar] [CrossRef]
- Jeelani, R.; Konwar, D.; Khan, A.; Kumar, D.; Chakraborty, D.; Brahma, B. Reassessment of temperature-humidity index for measuring heat stress in crossbred dairy cattle of a sub-tropical region. J. Therm. Biol. 2019, 82, 99–106. [Google Scholar] [CrossRef]
- Brode, P.; Fiala, D.; Blazejczyk, K.; Holmer, I.; Jendrizky, G.; Kampmann, B.; Tinz, B.; Havenith, G. Deriving the operational procedure for the universal thermal climatic index (UTCI). Int. J. Biomateorol. 2012, 56, 481–494. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I. Studies on genetics of heat tolerance in dairy cattle with reduced weather information via cluster analysis. J. Dairy Sci. 2002, 85, 1586–1589. [Google Scholar] [CrossRef]
- Bohmanova, J.; Misztal, I.; Cole, J.B. Temperature humidity indices as indicators of milk production losses due to heat stress. J. Dairy Sci. 2006, 90, 1947–1956. [Google Scholar] [CrossRef]
- Weber, J.A.; Park, S.G.; Luria, V.; Jeon, S.; Kim, H.M.; Jeon, Y.; Bhak, Y.; Jun, J.H.; Kim, S.W.; Hong, W.H.; et al. The whale shark genome reveals how genomic and physiological properties scale with body size. Proc. Natl. Acad. Sci. USA 2020, 117, 20662–20671. [Google Scholar] [CrossRef]
- Blache, D.; Maloney, S.K. New physiological measures of the biological cost of responding to challenges. In Advances in Sheep Welfare; Ferguson, D.M., Lee, C., Fisher, A., Eds.; Elsevier: London, UK, 2017; pp. 73–104. [Google Scholar]
- Gauly, M.; Ammer, S. Challenges for dairy cow production systems arising from climate changes. Animals 2020, 14, 196–203. [Google Scholar] [CrossRef]
- Yadav, B.; Singh, G.; Wankar, A. Acclimatization dynamics to extreme heat stress in crossbred cattle. Biol. Rhythm Res. 2021, 52, 524–534. [Google Scholar] [CrossRef]
- Sammad, A.; Wang, Y.J.; Umer, S.; Lirong, H.; Khan, I.; Khan, A.; Ahmad, B.; Wang, Y. Nutritional physiology and biochemistry of dairy cattle under the influence of heat stress: Consequences and opportunities. Animals 2020, 10, 793. [Google Scholar] [CrossRef]
- Archana, P.R.; Aleena, J.; Pragna, P.; Vidya, M.K.; Niyas, A.P.A.; Bagath, M.; Krishnan, G.; Manimaran, A.; Beena, V.; Kurien, E.K.; et al. Role of heat shock proteins in livestock adaptation to heat stress. J. Dairy Vet. Anim. Res. 2017, 5, 13–19. [Google Scholar]
- Murphy, K.T.; Petersen, A.C.; Goodman, C.; Gong, X.; Leppik, J.A.; Garnham, A.P.; Cameron-Smith, D.; Snow, R.J.; McKenna, M.J. Prolonged submaximal exercise induces isoform-specific Na+-K+-ATPase mRNA and protein responses in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, 414–424. [Google Scholar] [CrossRef][Green Version]
- Wyckelsma, V.L.; Perry, B.D.; Bangsbo, J.; McKenna, M.J. Inactivity and exercise training differentially regulate abundance of Na+-K+-ATPase in human skeletal muscle. J. Appl. Physiol. 2019, 127, 905–920. [Google Scholar] [CrossRef]
- Sahoo, S.S.; Mishra, C.; Rout, M.; Nayak, G.; Mohanty, S.T.; Panigrahy, K.K. Comparative in silico and protein-protein interaction network analysis of ATP1A1 gene. Gene Rep. 2016, 5, 134–139. [Google Scholar] [CrossRef]
- Signor, S.A.; Nuzhdin, S.V. The evolution of gene expression in cis and trans. Trends Genet. 2018, 34, 532–544. [Google Scholar] [CrossRef]
- Kaushik, R.; Goel, A.; Rout, P.K. Differential expression and characterization of ATP1A1 exon17 gene by high resolution melting analysis and RT-PCR in Indian goats. Mol. Biol. Rep. 2019, 46, 5273–5286. [Google Scholar] [CrossRef]
- Kaur, R.; Sharma, A.; Sodhi, M.; Swami, S.K.; Sharma, V.L.; Kumari, P.; Verma, P.; Mukesh, M. Sequence characterization of alpha 1 isoform (ATP1A1) of Na+/K+-ATPase gene and expression characteristics of its major isoforms across tissues of riverine buffaloes (Bubalus bubalis). Gene Rep. 2018, 10, 97–108. [Google Scholar] [CrossRef]
- Idris, M.; Uddin, J.; Sullivan, M.; McNeill, D.M.; Phillips, C.J. Non-invasive physiological indicators of heat stress in cattle. Animals 2021, 11, 71. [Google Scholar] [CrossRef]
- Moorjani, P.; Amorim, C.E.G.; Arndt, P.F.; Przeworski, M. Variation in the molecular clock of primates. Proc. Natl. Acad. Sci. USA 2016, 113, 10607–10612. [Google Scholar] [CrossRef]
- Loeza-Quintana, T.; Adamowicz, S.J. Iterative calibration: A novel approach for calibrating the molecular clock using complex geological events. J. Mol. Evol. 2018, 86, 118–137. [Google Scholar] [CrossRef]
- Di Marco, M.; Pacifici, M.; Maiorano, L.; Rondinini, C. Drivers of change in the realised climatic niche of terrestrial mammals. Ecography 2021, 44, 1–11. [Google Scholar] [CrossRef]
- Bulmer, M.; Wolfe, K.H.; Sharp, P.M. Synonymous nucleotide substitution rates in mammalian genes: Implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. USA 1991, 88, 5974–5978. [Google Scholar] [CrossRef]
- Mignone, F.; Gissi, C.; Liuni, S.; Pesole, G. Untranslated regions of mRNAs. Genome Biol. 2002, 3, 1–10. [Google Scholar] [CrossRef]
- Baker, E. mRNA stability and localization. In Lewin’s Genes X, 10th ed.; Lewin, B., Krebs, J.E., Goldstein, E.S., Kilpatrick, S.T., Eds.; Jones and Bartlett Learning: London, UK, 2011; pp. 618–639. [Google Scholar]
- Genuth, N.R.; Barna, M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Meyer, S.; Temme, C.; Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 2017, 51, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Tomanek, L. Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs. J. Exp. Biol. 2010, 213, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Fuller, A.; Mitchell, D.; Maloney, S.K.; Hetem, R.S.; Fonsêca, V.F.; Meyer, L.C.; van de Ven, T.M.; Snelling, E.P. How dryland mammals will respond to climate change: The effects of body size, heat load and a lack of food and water. J. Exp. Biol. 2021, 224, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Renaudeau, D.; Collin, A.; Yahav, S.; De Basilio, V.; Gourdine, J.L.; Collier, R.J. Adaptation to hot climate and strategies to alle viate heat stress in livestock production. Animals 2012, 6, 707–728. [Google Scholar]

| Confounding Variance | Studied Variance | ||
|---|---|---|---|
| Components | Inter Subject Variance | Processing Noise | Treatment Effect |
| Source | 1. Different base line expression 2. Different response to treatment | 1. Sampling 2. RT 3. qPCR | Difference between groups induced by treatment |
| Intervention | 1. Randomised 2. Used appropriate sample size 3. Used paired measures | 1. Used replicates 2. Normalized to reference gene or spike | Maximized effect (by selecting heat tolerant Vechur and Kasaragod and heat susceptible crossbred cattle |
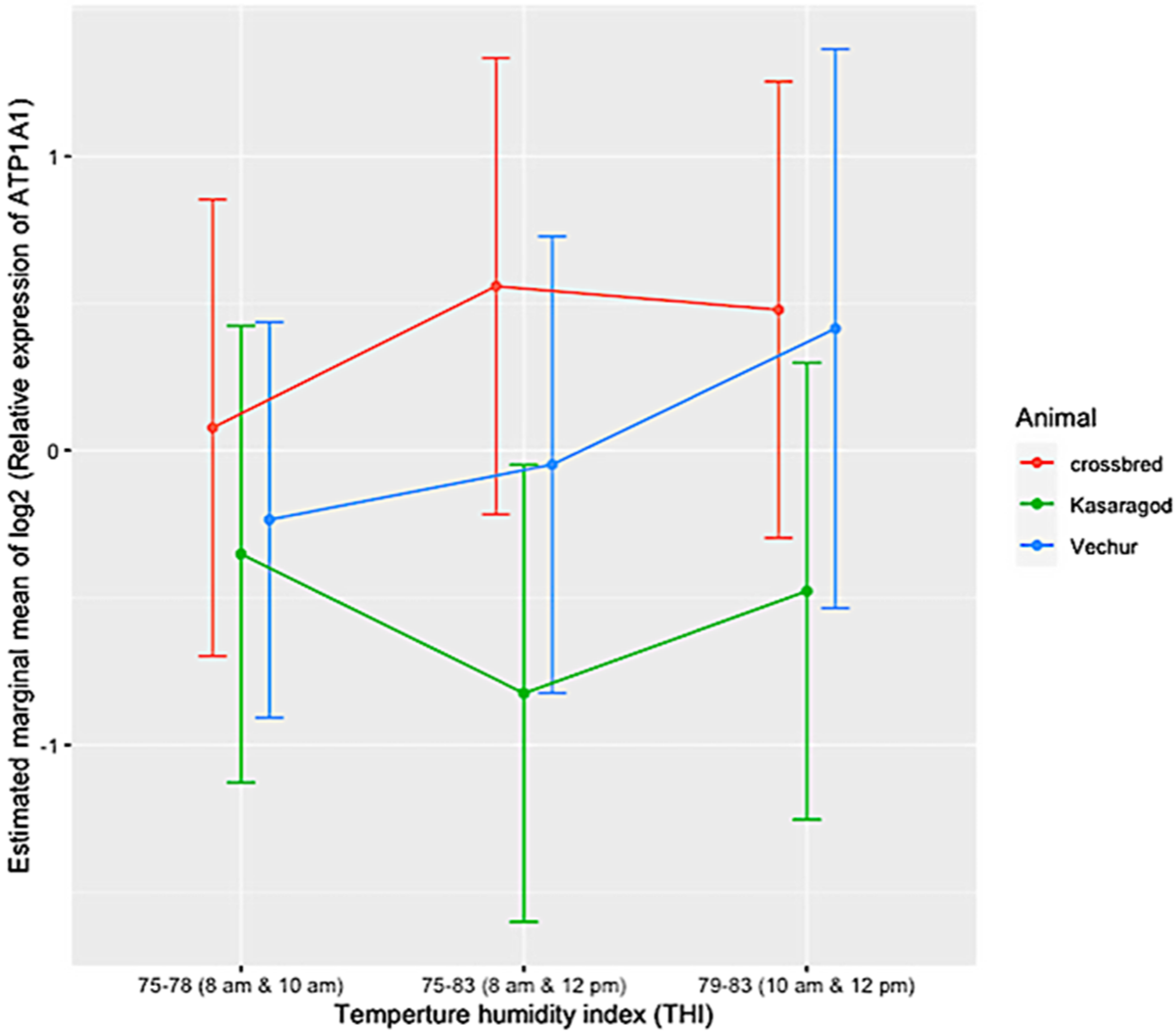
| Period | Breed | EMM | Lower CL | Upper CL |
|---|---|---|---|---|
| 1 | Vechur | −0.314 | −1.092 | 0.4641 |
| 2 | Vechur | 0.2764 | −0.502 | 1.0545 |
| 3 | Vechur | −0.0479 | −0.826 | 0.7302 |
| 1 | Kasaragod | −0.352 | −1.13 | 0.4261 |
| 2 | Kasaragod | −0.4776 | −1.256 | 0.3005 |
| 3 | Kasaragod | −0.8246 | −1.603 | −0.0465 |
| 1 | Crossbred | 0.0773 | −0.701 | 0.8554 |
| 2 | Crossbred | 0.4785 | −0.3 | 1.2566 |
| 3 | Crossbred | 0.5586 | −0.22 | 1.3367 |
| Exon | Genomic Coordinates | mRNA Coordinates | Length | Identity |
|---|---|---|---|---|
| Exon 1 | 918,643–918,655 | 1–13 | 13 | 100% |
| Exon 2 | 907,369–907,473 | 14–118 | 105 | 100% |
| Exon 3 | 906,689–906,748 | 119–178 | 60 | 100% |
| Exon 4 | 904,407–904,610 | 179–182 | 204 | 99.5% |
| Exon 5 | 903,621–903,734 | 383–496 | 114 | 100% |
| Exon 6 | 903,106–903,240 | 497–631 | 135 | 99.3% |
| Exon 7 | 902,879–902,996 | 632–749 | 118 | 97.5% |
| Exon 8 | 902,159–902,427 | 750–1018 | 269 | 98.5% |
| Exon 9 | 901,429–901,627 | 1019–1217 | 199 | 97% |
| Exon 10 | 901,068–901,177 | 1218–1327 | 110 | 99% |
| Exon 11 | 899,006–899,140 | 1328–1462 | 135 | 100% |
| Exon 12 | 898,152–898,344 | 1463–1655 | 193 | 99% |
| Exon 13 | 895,764–895,939 | 1656–1831 | 176 | 100% |
| Exon 14 | 894,642–894,778 | 1832–1968 | 137 | 100% |
| Exon 15 | 893,243–893,393 | 1969–2119 | 151 | 98.7% |
| Exon 16 | 892,478–892,648 | 2120–2288 | 169 | 100% |
| Exon 17 | 892,209–892,363 | 2289–2443 | 155 | 99.4% |
| Exon 18 | 891,714–891,837 | 2444–2567 | 124 | 98.4% |
| Exon 19 | 890,422–890,567 | 2568–2713 | 146 | 98.6% |
| Exon 20 | 890,166–890,296 | 2714–2844 | 131 | 100% |
| Exon 21 | 889,575–889,676 | 2845–2966 | 102 | 97.1% |
| Exon 22 | 887,420–887,511 | 2967–3038 | 92 | 98.9% |
| Exon 23 | 886,685–886,933 | 3039–3287 | 249 | 97.2% |
| Animal 1 | Animal 2 | Genetic Distance |
|---|---|---|
| KF286658.1 Bos t. indicus (Vechur) | NM 001076798.1 Bos t. taurus | 0.0076 |
| KF286658.1 Bos t. indicus (Vechur) | XM 027535865.1 Bos t. indicus × Bos t. taurus | 0.0081 |
| NM 001076798.1 Bos t. taurus | XM 027535865.1 Bos t. indicus × Bos t. taurus | 0.0004 |
| KF286658.1 Bos t. indicus (Vechur) | XM 005901993.2 Bos mutus | 0.0094 |
| NM 001076798.1 Bos t. taurus | XM 005901993.2 Bos mutus | 0.0024 |
| XM 027535865.1 Bos t. indicus × Bos t. taurus | XM 005901993.2 Bos mutus | 0.0024 |
| KF286658.1 Bos t. indicus (Vechur) | XM 010830730.1 Bison bison | 0.0093 |
| NM 001076798.1 Bos t. taurus | XM 010830730.1 Bison bison | 0.0239 |
| XM 027535865.1 Bos t. indicus × Bos t. taurus | XM 010830730.1 Bison bison | 0.0252 |
| KF286658.1 Bos t. indicus (Vechur) | XM 006058684.2 Bubalus bubalis | 0.0163 |
| NM 001076798.1 Bos t. taurus | XM 006058684.2 Bubalus bubalis | 0.0084 |
| XM 027535865.1 Bos t. indicus × Bos taurus | XM 006058684.2 Bubalus bubalis | 0.0082 |
| KF286658.1 Bos t. indicus (Vechur) | XM 018045956.1 Capra hircus | 0.0287 |
| NM 001076798.1 Bos t. taurus | XM 018045956.1 Capra hircus | 0.0208 |
| XM 027535865.1 Bos t. indicus × Bos t. taurus | XM 018045956.1 Capra hircus | 0.0211 |
| KF286658.1 Bos t. indicus (Vechur) | NM 001009360.1 Ovis aries | 0.0299 |
| NM 001076798.1 Bos t. taurus | NM 001009360.1 Ovis aries | 0.0210 |
| XM 027535865.1 Bos t. indicus × Bos t. taurus | NM 001009360.1 Ovis aries | 0.0214 |
| KF286658.1 Bos t. indicus (Vechur) | BC003077.2 Homo sapiens | 0.0877 |
| NM 001076798.1 Bos t. taurus | BC003077.2 Homo sapiens | 0.0832 |
| XM 027535865.1 Bos t. indicus × Bos t. taurus | BC003077.2 Homo sapiens | 0.0835 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elayadeth-Meethal, M.; Thazhathu Veettil, A.; Asaf, M.; Pramod, S.; Maloney, S.K.; Martin, G.B.; Rivero, M.J.; Sejian, V.; Naseef, P.P.; Kuruniyan, M.S.; et al. Comparative Expression Profiling and Sequence Characterization of ATP1A1 Gene Associated with Heat Tolerance in Tropically Adapted Cattle. Animals 2021, 11, 2368. https://doi.org/10.3390/ani11082368
Elayadeth-Meethal M, Thazhathu Veettil A, Asaf M, Pramod S, Maloney SK, Martin GB, Rivero MJ, Sejian V, Naseef PP, Kuruniyan MS, et al. Comparative Expression Profiling and Sequence Characterization of ATP1A1 Gene Associated with Heat Tolerance in Tropically Adapted Cattle. Animals. 2021; 11(8):2368. https://doi.org/10.3390/ani11082368
Chicago/Turabian StyleElayadeth-Meethal, Muhammed, Aravindakshan Thazhathu Veettil, Muhasin Asaf, Sathiamoorthy Pramod, Shane K. Maloney, Graeme B. Martin, M. Jordana Rivero, Veerasamy Sejian, Punnoth Poonkuzhi Naseef, Mohamed Saheer Kuruniyan, and et al. 2021. "Comparative Expression Profiling and Sequence Characterization of ATP1A1 Gene Associated with Heat Tolerance in Tropically Adapted Cattle" Animals 11, no. 8: 2368. https://doi.org/10.3390/ani11082368
APA StyleElayadeth-Meethal, M., Thazhathu Veettil, A., Asaf, M., Pramod, S., Maloney, S. K., Martin, G. B., Rivero, M. J., Sejian, V., Naseef, P. P., Kuruniyan, M. S., & Lee, M. R. F. (2021). Comparative Expression Profiling and Sequence Characterization of ATP1A1 Gene Associated with Heat Tolerance in Tropically Adapted Cattle. Animals, 11(8), 2368. https://doi.org/10.3390/ani11082368








