Simple Summary
An understanding of the way that animals respond to heat stress is key to the development of adaptation and mitigation strategies for a changing climate. The response of mammals to heat exposure involves changes at every level of organization from molecular and cellular to systemic and behavioral. The concert of events involves many genes and gene products. The Na+/K+ ATPase ∝1 (ATP1A1), a product of the ATP1A1 gene, is important for the response to heat because it determines the activity of the Na+/K+ pump that is ubiquitous in cell membranes. It was shown recently that ATP1A1 is important in combating the oxidative stress that a cell faces and that it modulates the Src signaling pathway that is involved in the response to many stressors. Vechur cattle (dwarf Bos taurus indicus) are well known for their adaptability to the tropical heat and humidity that persists in their native state of Kerala, India. We here analyze the comparative expression profile of the ATP1A1 gene in heat-tolerant Vechur and Kasaragod (another dwarf B. t. indicus) cattle and a heat-intolerant crossbreed (B. t. taurus × B. t. indicus) and characterize the sequence of ATP1A1 mRNA in the Vechur genotype. Environmental stress and heat tolerance were measured. Expression profiling indicated that ATP1A1 was differentially expressed in the phenotypically disparate cattle breeds. A molecular evolutionary genetic analysis revealed that the divergent origin of dwarf cattle was adaptive in response to heat stress and suggests the potential use of ATP1A1 as a marker for heat tolerance.
Abstract
Climate change is an imminent threat to livestock production. One adaptation strategy is selection for heat tolerance. While it is established that the ATP1A1 gene and its product play an important role in the response to many stressors, there has been no attempt to characterize the sequence or to perform expression profiling of the gene in production animals. We undertook a field experiment to compare the expression profiles of ATP1A1 in heat-tolerant Vechur and Kasaragod cattle (Bos taurus indicus) with the profile of a heat-susceptible crossbreed (B. t. taurus × B. t. indicus). The cattle were exposed to heat stress while on pasture in the hot summer season. The environmental stress was quantified using the temperature humidity index (THI), while the heat tolerance of each breed was assessed using a heat tolerance coefficient (HTC). The ATP1A1 mRNA of Vechur cattle was amplified from cDNA and sequenced. The HTC varied significantly between the breeds and with time-of-day (p < 0.01). The breed–time-of-day interaction was also significant (p < 0.01). The relative expression of ATP1A1 differed between heat-tolerant and heat-susceptible breeds (p = 0.02). The expression of ATP1A1 at 08:00, 10:00 and 12:00, and the breed–time-of-day interaction, were not significant. The nucleotide sequence of Vechur ATP1A1 showed 99% homology with the B. t. taurus sequence. The protein sequence showed 98% homology with B. t. taurus cattle and with B. grunniens (yak) and 97.7% homology with Ovis aries (sheep). A molecular clock analysis revealed evidence of divergent adaptive evolution of the ATP1A1 gene favoring climate resilience in Vechur cattle. These findings further our knowledge of the relationship between the ATP1A1 gene and heat tolerance in phenotypically incongruent animals. We propose that ATP1A1 could be used in marker assisted selection (MAS) for heat tolerance.
1. Introduction
Tolerance to high heat and humidity is a vital functional trait in species that evolved in adverse environments [1,2,3] and is mediated by molecular mechanisms that demonstrate the complex interplay of genes and environment [4,5]. In this context, the relationship between body size and tolerance to heat stress becomes critical [6] and, in southern India, is exemplified by the indigenous dwarf B. t. indicus cattle—the Vechur, the smallest cattle genotype in the world, and the slightly larger Kasaragod—that are mainly reared in the integrated farming systems that prevail in the region. Smaller size and tolerance to heat stress evolved in parallel as an adaptation to the tropical climate prevailing in their native habitat [6]. As a consequence, these genotypes are considered to be an important genetic resource for selection for climate resilience [7,8].
Heat stress results in plasma electrolyte imbalance that is caused mainly by oxidative stress, and the genes associated with stress response and protein repair are upregulated in stressed animals, whereas the genes associated with biosynthesis, metabolism, and body conformation are downregulated [9]. A key to the response is the heat shock proteins (HSP), the expression of which increases while the expression of non-HSPs decreases [10]. For these reasons, the differential expression of the genes regulating these processes is considered as a measure of cellular stress [11].
Of particular interest is the gene controlling Na+/K+ adenosine triphosphatase (Na+/K+ ATPase), a cell membrane protein that controls membrane permeability by coupling the transport of three Na+ ions outward and two K+ ions inward [12]. This coupling of ions across the plasma membrane is achieved using energy released from ATP hydrolysis [13,14,15]. The enzyme has three subunits, alpha, beta, and gamma, among which the alpha subunit is the largest and possesses the site for ATP hydrolysis. There are four alpha ATPases, ATP1A1, ATP1A2, ATP1A3, and ATP1A4, that code for α1, α2, α3, and α4 protein subunits, respectively. The expression of ATP1A1 is ubiquitous and is important in maintaining Na+ and K+ homeostasis across the plasma membrane of all tissues in the body, including blood cells [16].
Cell membrane permeability is affected by heat stress because there are changes in the electrochemical gradient that are mediated, at least partly, by changes in the expression of the Na+/K+ ATPase [15]. The major isoform of this enzyme, α1, encoded by the ATP1A1 gene, is a prominent non-heat shock protein that is associated with the response to heat stress [17]. Alternative splicing of ATP1A1 mediates the HSP-mediated heat shock response by maintaining the ATP balance [18,19]. Thus, ATP1A1 is considered a key non-HSP gene marker for tolerance to heat stress [16]. Indeed, the bovine ATP1A1 gene, with a coding sequence of 3065 nucleotides scattered over 23 exons, has polymorphisms that are associated with heat tolerance traits in dairy cattle [20,21]. Polymorphisms in ATP1A1 are associated with heat tolerance, disease susceptibility, and fertility in several breeds of dairy cattle, including Jersey crossbred, Sahiwal, Holstein, and Cholistani [22,23,24,25,26,27,28,29]. In beef cattle, heat stress is associated with changes in the expression of ATP1A1 [30,31,32].
Glucocorticoids increase Na+/K+ ATPase mRNA levels in skeletal muscles [33]. Buckard et al. (2015) analyzed the role of ATP1A1 in signal transduction involving cardiotonic binding induced signals through an Src signaling mediated pathway [34]. The cardiotonic steroids digoxin, ouabain, and bufarin act as specific inhibitors of ATP1A1 [35,36,37].
Sequence characterization, molecular evolutionary analysis, and molecular clock analysis enable the identification of the selective pressures and evolutionary history of particular taxa in relation to their environment [3]. In the genus Bos, functional traits such as body size and tolerance to stress and diseases evolved in parallel in response to the environmental adversities in which the breeds evolved [6]. The molecular clock accounts for the genetic distance between the taxa and translates distance to the time using calibration rate, measured in terms of rate of genetic change per unit of time [3].
Thus, advances in genome sequencing, genome-wide association studies, and evolutionary analysis enabled the unravelling of the mechanistic ways by which animals gain tolerance to adverse climatic conditions. The knowledge at the molecular level of a key candidate gene (ATP1A1) in cellular metabolism advances this knowledge and helps to select animals with high tolerance. However, although the ATP1A1 gene plays a key role in metabolism, growth, production, reproduction, and environmental adaptation of animals, the molecular genetics and the evolutionary basis of the role of ATP1A1 in animals with different body size and heat tolerance are not clearly understood. Hence, this study was designed: (1) to assess heat tolerance in Vechur, Kasaragod and crossbred cattle using a heat tolerance coefficient (HTC); (2) to compare the differential expression profile of ATP1A1 in heat stressed animals; (3) to sequence, characterize, and compare the ATP1A1 mRNA in Vechur cattle; and (4) to perform molecular evolutionary analysis to study the evolution of the ATP1A1 gene.
2. Materials and Methods
Ten adult female animals each of Vechur, Kasaragod, and crossbred cattle were selected for the study. The Vechur and the Kasaragod animals were obtained from the Kerala Veterinary and Animal Sciences University Vechur Conservation Unit, and the crossbred animals were obtained from the Kerala Veterinary and Animal Sciences University livestock farm, Mannuthy. All animals were previously exposed to heat challenge conditions because they grazed outside in summer. During the experiment, the animals were allowed to graze on hot summer days without any shade and were given a regular diet of concentrate feed, with drinking water ad libitum. The study was approved by the institutional animal ethics committee of the Kerala Veterinary and Animal Sciences University. The detailed experimental design is explained in a previous article [6].
Temperature and relative humidity data were obtained from an automatic weather station at the Kerala Agricultural University about four kilometers from the location of the experiment. The environmental stress was evaluated using the temperature humidity index (THI): THI = (1.8 × Ta + 32) − (0.55 − 0.0055 × RH) × (1.8 × Ta − 26), where Ta = atmospheric temperature (°C) and RH = percent relative humidity [38]. Respiratory rates (RR) and rectal temperatures (RT) were recorded at half hour intervals from 08:00 to 14:00 for ten days. The heat tolerance of the animals was assessed using a heat tolerance coefficient (HTC): HTC = RR/23 + RT/38 [16].
Jugular blood (5 mL) was sampled from each animal three times each day at 08:00, 10:00, and 12:00 using a vacutainer containing 5 mg EDTA as anticoagulant under aseptic conditions. The samples were immediately placed on ice and transported to the laboratory where RNA was extracted using GeneiPure RNA kits (Cat. No. KT-173, Genei, Bangalore, India). The quality of the RNA was checked using gel electrophoresis, and RNA was quantified using a spectrophotometer (Nanodrop ND-1000, Thermo-Scientific, Waltham, MA, USA). A DNase1 kit (Cat. No. AMP-D1, Sigma-Aldrich, St. Louis, MO, USA) was used to inhibit DNAse, followed by cDNA synthesis using kits (Cat. No. K1621, Fermentas, Waltham, MA, USA).
2.1. Quantitative Real Time PCR (q-RT PCR)
The q-RT PCR experiment was designed to test whether gene expression differed between Vechur, Kasaragod, and crossbred animals at various levels of heat stress (Table 1). Extended blood sample storage or delays in processing due to technical issues reduced ATP1A1 expression, indicating a need for fresh blood samples and speedy processing for optimal measures of ATP1A1 expression. A nested design (hierarchal or clustered) was followed in the present study, which consisted of three animals from each group, from which three treatment samples were collected and extracted. After the RT reaction, each sample was split into three q-RT PCR reactions for effective distribution of variance components [39]. To compare gene expression profiles under thermal stress, the selection of appropriate reference genes was important [40]. In our study, ß-actin was used as the reference gene, as it was stably expressed during thermal stress compared to other usual housekeeping genes GAPDH and 18S rRNA [40,41,42].

Table 1.
Distribution of variance components followed in the gene expression study [39].
The q-RT-PCR used custom synthesized primer pairs using the mRNA sequence NM_1076798.1- F- TCCTCATCGGCATCATTGTAGCCA; R- AGCCTCCAGGTTCTTCACTAAGCA to amplify a 122 bp amplicon with an annealing temperature of 60 °C. Gene expression was profiled using Illumina Eco® q-RT PCR with SYBR green. The profiling relied on the comparison of expression of ATP1A1 with expression of the reference gene (ß-actin) to calculate ΔCt; the expression of the same gene in treatment samples (10:00 and 12:00) was compared with expression in control (calibrator) samples (at 08:00) to calculate ΔΔCt. For calculation of the relative expression, the comparative Ct method, also known as the 2−∆∆Ct method, was used: ∆∆Ct = ∆Ct,sample − ∆Ct,calibrator (∆Ct = Ct value for the target gene − Ct value of the reference gene). The fold changes in expression of ATP1A1 gene after exposure to heat stress were estimated by comparison with values of the control animals (before exposure) using the above formula RQ = 2−∆∆Ct. For q-RT-PCR, we used Maxima SYBR Green master mix (Cat. No. K0221, Thermo Scientific).
Initially, the PCR conditions were optimized by using various concentrations of MgCl2 and dNTPs and by testing various time–temperature combinations for annealing and extension. The combination that gave the best result in terms of yield and specificity of the product was further used to amplify the samples for q-RT PCR and PCR in a thermal cycler (Bio-Rad, Thermal CyclerTM, Hercules, CA, USA). The PCR products were confirmed by gel electrophoresis using ethidium bromide staining in 2% agarose gel documented in a gel documentation system (Bio-Rad, Hercules, CA, USA).
For q-RT-PCR, separate PCR reactions were set up for ATP1A1 and ß-actin genes. First, a master mix was prepared for the required number of reactions. The Illumina Eco® q-RT PCR system utilized 48-well micro-plates for the reactions. The template (cDNA) was loaded separately into designated wells. Initial denaturation at 95 °C for 10 min was followed by denaturation at 95 °C for 15 s and repeated for 35 cycles. Data recording was done at the annealing stage set 60 °C for 60 s. The reaction specificity was assessed using melt curve analysis that consisted of denaturation at 95 °C for 15 s and annealing at 55 °C for 15 s followed by 95 °C for 15 s. The melt curve data were recorded in the denaturation step. The data were analyzed for relative quantification using the ΔΔCt method.
2.2. Amplification of the ATP1A1 Gene
The ATP1A1 mRNA of Vechur cattle was amplified from cDNA and sequenced. Primers were designed to amplify the ATP1A1 gene based on the mRNA sequence NM_1076798.1 using the primer blast tool at NCBI. The primers were checked for integrity using Sequence Manipulation Suite software (http://www.bioinformatics.org/sms2/html). The two primers selected (F-TATGGGGAAGGGGGTTGGACGTGATA; R-GTCGTTTCCACAGACGGATGTTTCTC) were custom-synthesized by Sigma-Aldrich.
The PCR conditions were optimized by using various concentrations of MgCl2 and dNTPs and by testing various time–temperature combinations for annealing and extension. JumpStart® Taq DNA polymerase (Cat. No. D-9307, Sigma-Aldrich) was used for amplification. The PCR was carried out in a volume of 50 µL in a 200 µL PCR tube. The combination that gave the best result in terms of yield and specificity of the product was further used for amplification of the samples, and PCR was carried out in a thermal cycler (Bio-Rad, Thermal CyclerTM, Hercules, CA, USA). The PCR conditions were as follows: initial denaturation at 96 °C for 30 s, followed by 35 cycles with denaturation at 94 °C for 15 s; primer-specific annealing temperature of 60 °C 30 s to specifically amplify the target region; extension at 68 °C for 3.5 min followed by final extension at 68 °C for 25 min. The PCR product was checked with 0.8% agarose gel prepared in 1X TAE buffer and incorporated with ethidium bromide along with a 1 Kb ladder (GeneRulerTM Cat. No. SM0311, ThermoFisher Scientific, Waltham, MA, USA). The gels were electrophoresed in 1X TAE buffer for 45 min at 75 V (5 V/cm) and documented in a gel documentation system (Bio-Rad, Hercules, CA, USA).
2.3. Sequence, Molecular Clock, and Data Analysis
The PCR-amplified ATP1A1 gene from Vechur cattle was sequenced and assembled commercially by Bioserve Biotechnologies (India) Pvt. Ltd. (Hyderabad, India) using the dideoxynucleotide chain termination sequencing method in an automated DNA sequencer (Applied Biosystems, ThermoFisher Scientific, Waltham, MA, USA). The raw sequence data were verified using the chromatogram to obtain clean sequence data. For comparison of similarity between the sequences, multiple sequence alignment feature of Clustal W2 was used to obtain the alignments, and a paired BLAST search was carried out for each pair to confirm paired sequence similarities. BLASTn analysis was done to obtain sequence similarity with nucleotide sequences available on Genbank. Most of these programs were in the CoreSuit 10 software DNASTAR Inc. (Madison, WI, USA), available at the bioinformatics laboratory of Department of Animal Breeding, Genetics and Biostatistics, College of Veterinary and Animal Sciences, Mannuthy, India.
The 3287 bp sequence obtained was analyzed for homology by BLAST search at the National Centre for Biotechnology Information (NCBI) site using BLAST program (http://www.ncbi.nlm.nih.gov/BLAST). The raw ATP1A1 sequences were annotated based on the bovine ATP1A1 (Accession # NM_ 001076798.1) gene. The Sequence Manipulation Suite was used for generating, formatting, and analyzing DNA and protein sequences. This program is available at http://www.bioinformatics.org/sms2/html. Multiple sequence alignment was done using the European Bioinformatics Institute (EBI) tool, Clustal (http://align.genome.jp/), and the SPLIGN program available at the NCBI site [43]. Using SPLIGN, the sequence was compared simultaneously with the sequence in the Bos t. taurus reference genome (1378962611).
For molecular clock analysis, the evolutionary history of the ATP1A1 gene was inferred using the neighbor-joining method [44]. The optimal tree with the sum of branch length = 0.12680380 was selected. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was shown next to the branches [45]. The evolutionary distances were computed using the maximum composite likelihood method and are presented in the units of the number of base substitutions per site [46]. This analysis involved 9 nucleotide sequences. Codon positions included were 1st+2nd+3rd+noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 3871 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [47,48].
Subsequently, a timetree was inferred by applying the RelTime method in the constructed phylogenetic tree [49,50]. The branch lengths were calculated using the maximum likelihood (ML) method and the Tamura-Nei substitution model [51]. The timetree was computed using 1 calibration constraint—the split of B. t. taurus and B. t. indicus. The method of Tao et al. (2019) was used to set minimum and maximum time boundaries on nodes for which calibration densities were provided [52]. The estimated log likelihood value of the tree was −8754.60. The human ATP1A1 nucleotide sequence was taken as an outgroup. Codon positions included were 1st+2nd+3rd+noncoding.
The statistical analysis was done in R (Version 4.1.0) [53]. Values for HTC and relative expression were log-transformed before analysis of variance. The effects of breed, period, and period–breed interaction was estimated. Estimated marginal means are presented. The effect of THI on HTC was also compared among animal groups. Pair-wise comparison was done using the TukeyHSD test in R. The level of significance was set at p < 0.05.
3. Results
The ambient temperature and humidity increased progressively from 08:00 to 14:00. The maximum air temperature during the study period was 31.1 ° C, the relative humidity was 83.8%, the wind speed was 5.8 m/s, and the solar radiation was 1.3 MJ. The calculated THI increased from 75 at 08:00 to 83 at 14:00. There was significant variation in HTC between the groups (p < 0.01). The effects of period and period–breed interaction was also significant (p < 0.01) (Figure 1 and Figure 2). Compared to dwarf cattle, the HTC increased in crossbred animals when the THI increased (Figure 1 and Figure 2). The estimated marginal means (emm) of ATP1A1 relative expression are given in Table 2. The expression profile of ATP1A1 varied significantly between the breeds (p = 0.02). However, the period and the period–breed interactions were not significant for ATP1A1 expression (Figure 3). Pair-wise comparison of ATP1A1 expression levels in different lineages showed significant difference in expression between Kasargode and crossbred cattle (p = 0.009).
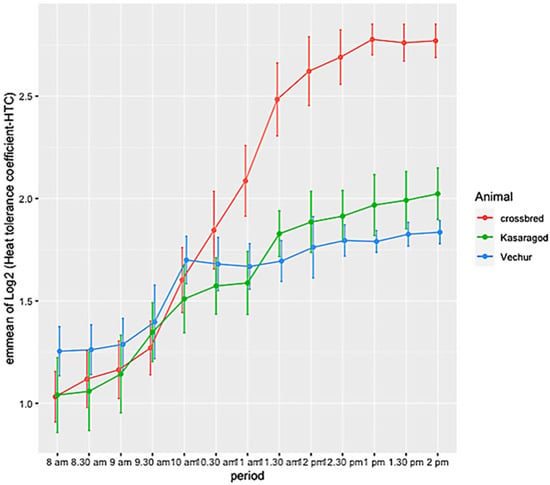
Figure 1.
Estimated marginal means (emmean) of the heat tolerance coefficient (HTC) in crossbred, Kasaragod, and Vechur cattle, from 08:00 to 14:00.
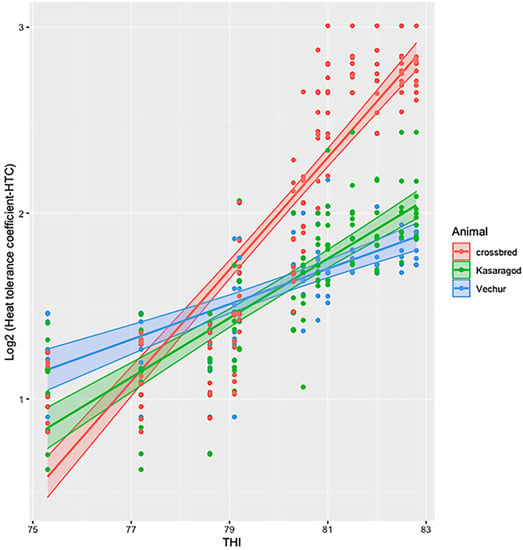
Figure 2.
Estimated marginal means of heat tolerance coefficient (HTC) in crossbred, Kasaragod, and Vechur cattle at different THI levels.

Table 2.
Estimated marginal means (EMM) of the relative expression of ATP1A1 gene in different breeds of cattle at different periods from 08:00 to 12:00 (standard error = 0.37; df = 18). Period 1 was between 08:00 and 10:00; period 2 was between 10:00 and 12:00; period 3 was between 08:00 and 12:00.
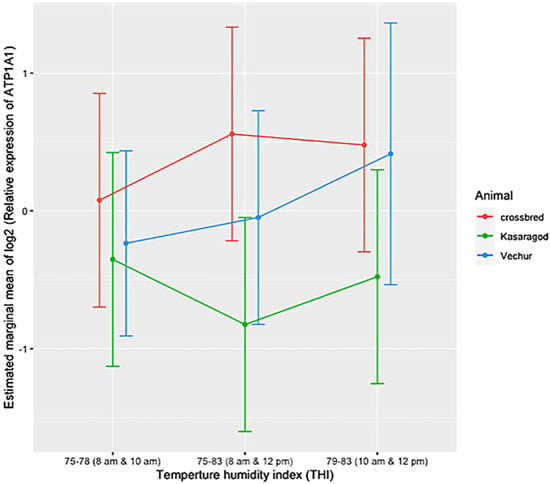
Figure 3.
Relative expression of the ATP1A1 gene in Vechur, Kasaragod, and crossbred cattle during different periods from 08:00 h to 12:00 h. Period 1 was between 08:00 and 10:00; period 2 was between 10:00 and 12:00; period 3 was between 08:00 and 12:00. The estimated marginal mean is shown on the y axis. The expression profile of ATP1A1 varied significantly among the breeds (p = 0.02). However, the effects of time and the time x breed interaction were not significant. Pair-wise comparison of ATP1A1 expression levels in different lineages showed significant difference in expression between Kasargode and crossbred cattle (p = 0.009).
The gel image of 3287 bp PCR amplified ATP1A1 gene is given in Figure S1. The sequence is available in NCBI (Accession number KF286658.1). Details of genomic structure and coordinates of ATP1A1 in Vechur are given in Table 3. A total of 36 variations in the ATP1A1 gene of Vechur were detected when compared with Bos t. taurus (Figures S2–S4). The percent identity and the divergence of the ATP1A1 gene and protein are given in Figure 4 and Figure 5. The protein sequence showed 98% homology with B. t. taurus cattle and B. grunniens (yak) and 97.7% with Ovis aries (sheep). Pair-wise genetic distances between the taxons are given in Table 4. The calculated genetic distances between Vechur, B.t. taurus, and B.t. taurus × B. t. indicus were 0.0076 and 0.0081, respectively. The evolutionary history of the ATP1A1 gene in Vechur cattle, inferred using the neighbor-joining method, is given in Figure S5. The optimal tree with the sum of branch length = 0.12680380 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The timetree depicting the evolutionary molecular clock of the ATP1A1 gene in dwarf Vechur (B. t. indicus) cattle is given in Figure 6. The phylogenetic tree shown in Figure S5 was used for molecular clock analysis. The branch lengths were calculated using the maximum likelihood (ML) method and the Tamura-Nei substitution model inferred by applying the RelTime method. Molecular clock analysis revealed the ancient origin of Vechur cattle compared to Bos t. taurus (Figure 6).

Table 3.
Details of genomic structure and coordinates of ATP1A1.
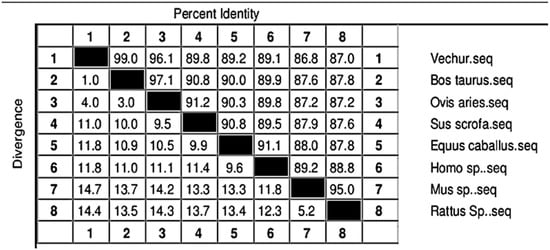
Figure 4.
Percent identity (upper half) and divergence (lower half) of the ATP1A1 gene of Vechur cattle with similar sequences in the database.
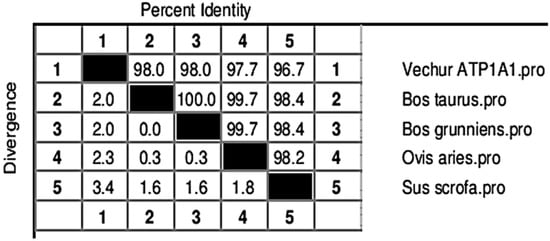
Figure 5.
Percent identity (upper half) and divergence (lower half) of the predicted ATP1A1 protein sequence of Vechur cattle with similar sequences in the database.

Table 4.
Pair-wise genetic distance between different lineages based on the mRNA sequence comparison of the ATP1A1 gene.
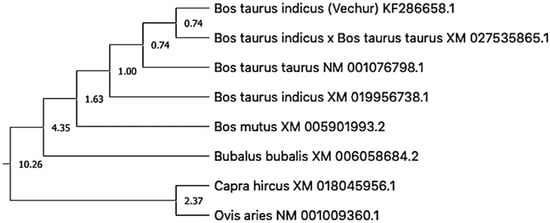
Figure 6.
Timetree depicting the evolutionary molecular clock of the ATP1A1 gene in dwarf Vechur (Bos taurus indicus) cattle. The branch lengths were calculated using the maximum likelihood (ML) method and the Tamura-Nei substitution model inferred by applying the RelTime method.
4. Discussion
Under thermoneutral conditions, all three groups of animals showed similar physiological responses, as indicated by the HTC. However, as the heat load increased, the HTC became higher in the crossbred cattle than in the dwarf Vechur and Kasaragod animals. Using expression profiling, we found that the expression of the ATP1A1 gene significantly varied between the heat-tolerant dwarf Vechur and Kasaragod cattle and the heat stress susceptible, standard sized, crossbred cattle. However, when THI increased, among the dwarf cattle lineages (Vechur and Kasaragod), Vechur had a higher level of ATP1A1 expression compared to Kasaragod animals. Although superior thermotolerance of dwarf cattle was evident in their physiological responses, the differential ATP1A1 expression upon increasing heat load among dwarf lineages requires further investigation.
The molecular clock analysis revealed separate clustering of Vechur cattle along with Bison and B. taurus cattle. Previous molecular clock analysis revealed that taurine and indicine cattle diverged around 1 mya [54,55]. The rate of DNA evolution and, hence, the molecular clock are primarily affected by body size and temperature [56,57,58]. The smaller body size of Vechur cattle might favor faster adaptive evolution to high heat and humidity in their lineage [59].
The experiment was conducted from 08:00 to 14:00, as the physiological responses were found to be maximum at 14:00 and minimum at 02:00 during all seasons [60]. The THI values reached a maximum in the afternoon, and the minimum was early in the morning [61]. Cattle are generally considered stressed above a THI of 72 [62]. The THI is similar to UTCI (universal thermal climate index) that is used in humans [63]. It can be used to account for the effect of heat stress on production of dairy cattle [64]. The THI is used as an indicator of milk production losses due to heat stress, and indices with larger weights on humidity are more suitable for humid climates [65]. The THI threshold refers to a value at which detectable change begins in the animal, denoted by THIon. The THI does not include important climatic variables such as solar radiation and wind speed. Likewise, it does not include management factors (the effect of shade) or animal factors (genotype differences).
Climate resilience is related to metabolic efficiency and body size of the organism, as physiological and genomic properties scale with body size [66]. Species that are less metabolically efficient have a larger metabolic heat load to deal with, which becomes difficult when the environment is not conducive to heat loss by dry or evaporative routes, such as hot and humid conditions exemplified by THI higher than 72 [67]. More metabolically efficient species/breeds have the potential to tolerate acute and chronic climate stress [68,69,70]. Heat shock proteins (HSPs) are studied in detail in relation to the enhanced thermotolerance [71]. Due to low specificity of HSPs, other non-HSPs were suggested as markers of thermotolerance. Hence, non-heat shock proteins such as ATP1A1 are considered candidate genes for thermotolerance and suggested as markers for selection for heat tolerance, disease resistance, low carbon foot print, conservation, and sustainable intensification [10,16]. To assess the impact of climate change on animals, several genetic markers were suggested [10]. Na+/K+ ATPase is the major class of enzymes that are involved in the membrane permeability of cells. An increased expression of ATP1A1 mRNA during prolonged exercise was reported without a corresponding increase in protein levels [72]. ATP1A1 is extensively studied in exercise physiology and inactivity in humans [73]. ATP1A1 interacts with other proteins and modulates hypertension and feed intake [74].
During thermal stress, multiple gene networks are activated [75]. Thus, the changes in ATP1A1 expression that we observed in the present study may have been influenced by many factors such as increased expression of heat shock factors and heat shock proteins, altered metabolism of glucose, amino acids, and fat, and altered immune responses [10]. Previous studies revealed breed specific nucleotide diversity and expression patterns of ATP1A1 in Indian goats and buffaloes [76,77]. Hence, ATP1A1 could be used as a marker for selection for heat tolerance [70].
Molecular clock theory provides a way to estimate variation in the rate of molecular evolution [78,79]. Several methods can be used to calibrate the molecular clock, including complex geological events [80]. In the case of vertebrates, body size and temperature significantly affect the molecular clock and thus in combination influence metabolism and the rate of evolution [56,57,58]. Species with a smaller dispersal range and large body mass tend to experience niche shrink in response to environmental perturbations and may require adaptive or plastic tolerance to survive in a warming climate [81]. The relative synonymous substitution rate after adjusting for taxa-specific and G+C constituent influences is fast for ATP1A1 [82]. Vechur, being the smallest breed of cattle, compared with standard size crossbred cattle make an ideal model for the evolutionary molecular clock analysis of large ruminants.
The sequence data revealed differences in the mRNA sequence of the ATP1A1 gene in Vechur cattle compared to B. t. taurus cattle, which was possibly one of the reasons for altered expression of these genes in Vechur cattle during thermoneutral condition. The 5′ and the 3′ UTRs of eukaryotic mRNAs were shown to influence several aspects of gene expression [83]. The 3′ UTR influences mRNA stability and translation initiation and, hence, mutations in the 3′ UTR are associated with a number of diseases [84].
Selection for heat stress is possible and particularly effective for environments with a high average THI. Continued selection for production ignoring heat tolerance results in a decrease in heat tolerance, as the genetic correlation between general production and heat tolerance is around −0.3. Thus, a combined selection for production and heat tolerance is possible [64]. For crossbreeds, the ability to acclimate is limited. The findings of the current study suggest that crossbreeds maximized their molecular safety factors, as inferred from the higher levels of expression of ATP1A1 at thermoneutral conditions compared to dwarf cattle, which does not allow for further adjustments to current changes in climate [85,86,87,88]. These animals are living close to their stress tolerance limit, and a global warming scenario is likely to push them beyond that which may ultimately lead to further decrease in productivity [89,90].
Thus, the first time, we have provided a quantitative fingerprint of expression of ATP1A1 genes in thermotolerant Vechur and Kasaragod cattle compared to thermally sensitive crossbred cattle under thermoneutral and heat stress conditions. The method is reproducible and sensitive enough to detect different levels of expression of ATP1A1 in different thermal conditions. The results have implications for cattle in a global warming scenario, as alleles for thermotolerance can be detected by allele mining or expression QTL (eQTL) to develop thermotolerant breeds in future [89,90].
5. Conclusions
The performance of livestock is evaluated in three key areas: growth, production, and reproduction. To maintain thermal equilibrium, cattle rely on a delicate balance of heat production and heat loss. Mechanisms of adaptation include morphological, behavioral, biochemical, and physiological changes, and the capacity of animals to adapt is limited primarily by their genetic makeup. The stress response is influenced by a number of factors including species, breed, age, sex, previous exposure to stress, health status, performance, and body condition. Subsequent acclimatization may alleviate the strain that develops in response to stress, but performance may not return to pre-stress levels. In the present study, it was found that ATP1A1 expression was significantly associated with heat tolerance in different populations studied. Additionally, we emphasize the potential use of ATP1A1 as a marker for marker assisted selection (MAS) for heat tolerance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani11082368/s1, Additional materials and methods, Figure S1: Gel demonstrating the 3287 bp PCR amplified product of ATP1A1 gene of Vechur cattle in 1% agarose gel, Figure S2: Results of mRNA (cDNA) to genomic alignment created by Splign. Graphical view the alignment between Vechur ATP1A1 sequence KF 286658.1 and Bos taurus reference genome (1378962611). Figure S3: Tabular format for the alignment between Vechur ATP1A1 sequence KF 286658.1 and Bos taurus reference genome (1378962611). Figure S4: Segments and boundaries of the aligned regions of 23 exons between Vechur ATP1A1 sequence (KF 286658.1) and Bos taurus reference genome (1378962611). Figure S5: The evolutionary history of ATP1A1 gene in Vechur cattle inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 0.12680380 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.
Author Contributions
Conceptualization, M.E.-M., S.K.M., G.B.M., V.S. and M.R.F.L.; Data curation, M.E.-M., M.A., P.P.N. and M.S.K.; Formal analysis, M.E.-M., M.A., P.P.N. and M.S.K.; Funding acquisition, M.E.-M. and M.R.F.L.; Investigation, M.E.-M., A.T.V., S.P., S.K.M., P.P.N. and M.S.K.; Methodology, A.T.V., S.P., S.K.M., M.J.R. and M.R.F.L.; Project administration, M.E.-M., A.T.V., S.P., S.K.M., M.J.R. and M.R.F.L.; Resources, A.T.V., G.B.M. and M.J.R.; Software, P.P.N. and M.S.K.; Supervision, A.T.V., M.J.R., P.P.N. and M.R.F.L.; Validation, M.E.-M.; Visualization, M.E.-M., M.A., M.J.R., V.S. and M.S.K.; Writing—original draft, M.E.-M., S.K.M., G.B.M., M.J.R., V.S. and M.R.F.L.; Writing—review and editing, M.A., S.K.M., M.J.R., V.S., P.P.N. and M.S.K. All authors have read and agreed to the published version of the manuscript.
Funding
Muhammed Elayadeth-Meethal was the recipient of Rothamsted International Fellowship and worked within the Soil to Nutrition Institute Strategic Research Programme (BBS/E/C/00010300) under Tom H. Misselbrook and MRFL. We also thank The Crawford Fund, Australia, for providing fellowship to MEM for training under SKM at UWA on measurements in thermal physiology (Project No. RA/3/100/1246). The work was done as part of PhD work of MEM. The Department of Animal Husbandry, Government of Kerala, India deputed MEM for the PhD.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee of the College of Veterinary and Animal Sciences, Mannuthy, Thrissur, Kerala, India (No. 6554/2012 dated 29 February 2013).
Data Availability Statement
ATP1A1 mRNA sequence of Vechur cattle is available at NCBI (accession number KF286658.1).
Acknowledgments
Kerala Veterinary and Animal Sciences University, Kerala, India (KVASU), University of Western Australia, Perth, Australia (UWA) and Rothamsted Research, UK (RReS) are all members of Global Farm Platform initiative (https://www.globalfarmplatform.org/ that attracts researchers from different communities and disciplines seeking to develop sustainable ruminant production globally. We thank KVASU, UWA, and RReS, North Wyke, Devon, for providing facilities for the study. The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia for funding this work through Research Group Program under Grant No: RGP 2/191/42.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Eisler, M.C.; Lee, M.R.F.; Tarlton, J.F.; Martin, G.B.; Beddington, J.; Dungait, J.A.; Greathead, H.; Liu, J.; Mathew, S.; Miller, H.; et al. Agriculture: Steps to sustainable livestock. Nature 2014, 507, 32–34. [Google Scholar] [CrossRef]
- Van Zanten, H.H.; Herrero, M.; Van Hal, O.; Roos, E.; Muller, A.; Garnett, T.; Gerber, P.J.; Schader, C.; De Boer, I.J. Defining a land boundary for sustainable livestock consumption. Glob. Chang. Biol. 2018, 24, 4185–4194. [Google Scholar] [CrossRef] [PubMed]
- Bromham, L.; Penny, D. The modern molecular clock. Nat. Rev. Genet. 2003, 4, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Huang, B.; Zhan, J.; Wang, J.; Qu, K.; Zhang, F.; Shen, J.; Jia, P.; Ning, Q.; Zhang, J.; et al. Whole-genome analyses identify loci and selective signals associated with body size in cattle. J. Anim. Sci. 2020, 98, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Perini, F.; Cendron, F.; Rovelli, G.; Castellini, C.; Cassandro, M.; Lasagna, E. Emerging genetic tools to investigate molecular pathways related to heat stress in chickens: A review. Animals 2021, 11, 1–19. [Google Scholar]
- Elayadeth-Meethal, M.; ThazhathuVeettil, A.; Maloney, S.K.; Hawkins, N.; Misselbrook, T.H.; Sejian, V.; Rivero, M.J.; Lee, M.R. Size does matter: Parallel evolution of adaptive thermal tolerance and body size facilitates adaptation to climate change in domestic cattle. Ecol. Evol. 2018, 8, 10608–10620. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.Z.; Sasazaki, S.; Mannen, H. Genetic diversity and structure in Bos taurus and Bos indicus populations analyzed by SNP markers. Anim. Sci. 2010, 81, 281–289. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Kemper, K.E.; Hayes, B.J.; Zhang, Y.; Tier, B.; Barendse, W.; Reverter, A.; Goddard, M.E. Detection of quantitative trait loci in Bos indicus and Bos taurus cattle using genome-wide association studies. Genet. Sel. Evol. 2013, 45, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; Stiening, C.M.; Pollard, B.C.; Vanbaale, M.J.; Baumgard, L.H.; Gentry, P.C.; Coussens, P.M. Use of expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J. Anim. Sci. 2006, 84, 1–13. [Google Scholar] [CrossRef]
- Collier, R.J.; Collier, J.L.; Rhoads, R.P.; Baumgard, L.H. Genes involved in the bovine heat stress response. J. Dairy Sci. 2008, 91, 445–454. [Google Scholar] [CrossRef]
- Agnew, L.; Colditz, I.G. Development of a method of measuring cellular stress in cattle and sheep. Vet. Immunl. Immunopathol. 2008, 123, 197–204. [Google Scholar] [CrossRef]
- Rajamanickam, G.D.; Kastelic, J.P.; Thundathil, J.C. Na/K-ATPase regulates bovine sperm capacitation through raft-and non-raft-mediated signaling mechanisms. Mol. Reprod. Dev. 2017, 84, 1168–1182. [Google Scholar] [CrossRef]
- Mavrogonatou, E.; Papadimitriou, K.; Urban, J.P.; Papadopoulos, V.; Kletsas, D. Deficiency in the α1 subunit of Na+/K+-ATPase enhances the anti-proliferative effect of high osmolality in nucleus pulposus intervertebral disc cells. J. Cell. Physiol. 2015, 230, 3037–3048. [Google Scholar] [CrossRef]
- Liu, Y.X.; Xu, C.H.; Gao, T.Y.; Sun, Y. Polymorphisms of the ATP1A1 gene associated with mastitis in dairy cattle. Genet. Mol. Res. 2012, 11, 651–660. [Google Scholar] [CrossRef]
- Geering, K.; Kraehenbuhl, J.P.; Rossier, B.C. Maturation of the catalytic alpha-subunit of Na, K- ATPase during intracellular transport. J. Cell Biol. 1987, 105, 2613–2619. [Google Scholar] [CrossRef]
- Das, R.; Gupta, I.D.; Verma, A.; Singh, S.; Chaudhari, M.V.; Sailo, L.; Verma, N.; Kumar, R. Single nucleotide polymorphisms in ATP1A1gene and their association with thermotolerance traits in Sahiwal and Karan Fries cattle. India. J. Anim. Res. 2017, 51, 70–74. [Google Scholar]
- Wang, Z.; Huang, J.; Zhong, J.; Wang, G. Tissue specific alternative splicing and expression of ATP1B2 gene. Afr. J. Biotech. 2012, 39, 9485–9495. [Google Scholar]
- Wang, S.; Edens, F.W. Involvement of steroid hormones, corticosterone and testosterone, in synthesis of heat shock proteins in broiler chicken. Int. J. Poult. Sci. 2008, 7, 783–797. [Google Scholar] [CrossRef]
- Wang, L.; Schumann, U.; Liu, Y.; Prokopchuk, O.; Steinacker, J.M. Heat shock protein 70 (Hsp70) inhibits oxidative phosphorylation and compensates ATP balance through enhanced glycolytic activity. J. Appl. Physiol. 2012, 113, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S.; et al. Whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Zhou, X.; Li, D.Q.; Cui, Q.W.; Wang, G.L. Association of ATP 1 A 1 gene polymorphism with heat tolerance traits in dairy cattle. Genet. Mol. Res. 2010, 9, 891–896. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Li, H.; Zhou, X.; Wang, G. A Novel SNP of the ATP1A1 gene is associated with heat tolerance traits in dairy cows. Mol. Biol. Rep. 2011, 38, 83–88. [Google Scholar] [CrossRef]
- Vague, P.; Dufayet, D.; Coste, T.; Moriscot, C.; Jannot, M.F.; Raccah, D. Association of diabetic neuropathy with Na/K ATPase gene polymorphism. Diabetologia 1997, 40, 506–511. [Google Scholar] [CrossRef]
- Schlingmann, K.P.; Bandulik, S.; Mammen, C.; Tarailo-Graovac, M.; Holm, R.; Baumann, M.; Konig, J.; Lee, J.J.; Drogemoller, B.; Imminger, K.; et al. Germline de novo mutations in ATP1A1 cause renal hypomagnesemia, refractory seizures, and intellectual disability. Am. J. Hum. Genet. 2018, 103, 808–816. [Google Scholar] [CrossRef]
- Stregapede, F.; Travaglini, L.; Rebelo, A.P.; Cintra, V.P.; Bellacchio, E.; Bosco, L.; Alfieri, P.; Pro, S.; Zuchner, S.; Bertini, E.; et al. Hereditary spastic paraplegia is a novel phenotype for germline de novo ATP1A1 mutation. Clin. Genet. 2020, 97, 521–526. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.P.; Sanchez, G.; Mitra, A.; Numata, S.; Liu, L.C.; Blanco, G. Na, K-ATPase α4, and not Na, K-ATPase α1, is the main contributor to sperm motility, but its high ouabain binding affinity site is not required for male fertility in mice. J. Membr. Biol. 2021, 1–13. [Google Scholar]
- Das, R.; Gupta, I.D.; Verma, A.; Singh, A.; Chaudhari, M.V.; Sailo, L.; Upadhyay, R.C.; Goswami, J. Genetic polymorphisms in ATP1A1 gene and their association with heat tolerance in Jersey crossbred cows. India. J. Dairy Sci. 2015, 68, 50–54. [Google Scholar]
- Ramendra, D.; Gupta, I.D.; Archana, V.; Chaudhari, M.V.; Lalrengpuii, S.; Sohanvir, S. Identification of SNPs in ATP1A1 gene and their association with heat tolerance in Sahiwal and Karan Fries (Bos taurus× Bos indicus) cattle under tropical climatic condition. Indian, J. Dairy Sci. 2018, 71, 409–415. [Google Scholar]
- Imran, S.; Khan, M.S.; Qureshi, Z.I. Genetic characterization of Cholistani breed of cattle for ATP1A1 gene and its association to heat tolerance traits. Pak. J. Agric. Sci. 2021, 58, 229–234. [Google Scholar]
- Faheem, M.S.; Ghanem, N.; Gad, A.; Procházka, R.; Dessouki, S.M. Adaptive and biological responses of buffalo granulosa cells exposed to heat stress under In vitro condition. Animals 2021, 11, 794. [Google Scholar] [CrossRef]
- Kashyap, N.; Kumar, P.; Deshmukh, B.; Bhat, S.; Kumar, A.; Chauhan, A.; Bhushan, B.; Singh, G.; Sharma, D. Association of ATP1A1 gene polymorphism with thermotolerance in Tharparkar and Vrindavani cattle. Vet. World 2015, 8, 892–897. [Google Scholar] [CrossRef][Green Version]
- Chang, J.T.; Lowery, L.A.; Sive, H. Multiple roles for the Na, K-ATPase subunits Atp1a1 and Fxyd1, during brain ventricle development. Dev. Biol. 2012, 368, 312–322. [Google Scholar] [CrossRef]
- Thompson, C.B.; Dorup, I.; Ahn, J.; Leong, P.K.; McDonough, A.A. Glucocorticoids increase sodium pump α2-and β1-subunit abundance and mRNA in rat skeletal muscle. Am. J. Physiol. Cell Physiol. 2001, 280, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Burkard, C.; Verheije, M.H.; Haagmans, B.L.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J.; de Haan, C.A. ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J. Virol. 2015, 89, 4434–4448. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.H.; Ellakwa, D.E.S. SUMO pathway, blood coagulation and oxidative stress in SARS-CoV-2 infection. Biochem. Biophys. Rep. 2021, 26, 1–6. [Google Scholar]
- Medina-Ortiz, K.; Lopez-Alvarez, D.; Navia, F.; Hansen, T.; Fierro, L.; Castano, S. Identification of Na+/K+-ATPase α/β isoforms in Rhinella marina tissues by RNAseq and a molecular docking approach at the protein level to evaluate α isoform affinities for bufadienolides. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2021, 254, 1–14. [Google Scholar]
- Yang, N.; Shen, H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef]
- Bohmanova., J.; Misztal, I.; Tsuruta, S.; Norman, H.D.; Lawlor, T.J. Genotype by environment interaction due to heat stress. J. Dairy Sci. 2008, 91, 840–846. [Google Scholar] [CrossRef]
- Tichopad, A.; Kitchen, R.; Riedmaier, I.; Becker, C.; Kubista, M. Design and optimization of reverse transcription quantitative PCR experiments. Clin. Chem. 2009, 55, 1816–1823. [Google Scholar] [CrossRef]
- Xiang-Hong, J.; Hong, Y.Y.; Jin, X.H.; Mei, X.Y.; Rong, J.P.; Ming, L. Selection of reference genes for gene expression studies in PBMC from Bama miniature pig under heat stress. Vet. Immunol. Immunopathol. 2011, 144, 160–166. [Google Scholar] [CrossRef]
- Mahadav, A.; Kontsedelov, S.; Czosnek, H.; Ghanim, M. Thermotolerance and gene expression following heat stress in the whitefly Bamesia tobaci B and Q biotypes. Insect Biochem. Mol. Biol. 2009, 39, 668–676. [Google Scholar] [CrossRef]
- Eitam, H.; Vaya, J.; Brosh, A.; Orlow, A.; Khatib, S.; Izhaki, I.; Shabtay, A. Differential stress responses among newly received calves: Variations in reductant capacity and HSP gene expression. Cell Stress Chaperones 2010, 15, 865–876. [Google Scholar] [CrossRef]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Biol. Direct 2008, 3, 1–13. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef]
- Tamura, K.; Qiqing, T.; Kumar, S. Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol. Biol. Evol. 2018, 35, 1770–1782. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Tao, Q.; Tamura, K.; Mello, B.; Kumar, S. Confidence intervals for RelTime estimates of divergence times from molecular data. Mol. Biol. Evol. 2020, 37, 280–290. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Loftus, R.T.; MacHugh, D.E.; Bradley, D.G.; Sharp, P.M.; Cunningham, P. Evidence for two independent domestications of cattle. Proc. Natl. Acad. Sci. USA 1994, 91, 2757–2761. [Google Scholar] [CrossRef]
- MacHugh, D.E.; Shriver, M.D.; Loftus, R.T.; Cunningham, P.; Bradley, D.G. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 1997, 146, 1071–1086. [Google Scholar] [CrossRef]
- Gillooly, J.F.; Allen, A.P.; West, G.B.; Brown, J.H. The rate of DNA evolution: Effects of body size and temperature on the molecular clock. Proc. Natl. Acad. Sci. USA 2005, 102, 140–145. [Google Scholar] [CrossRef]
- Thomas, J.A.; Welch, J.J.; Woolfit, M.; Bromham, L. There is no universal molecular clock for invertebrates, but rate variation does not scale with body size. Proc. Natl. Acad. Sci. USA 2006, 103, 7366–7371. [Google Scholar] [CrossRef] [PubMed]
- Pulquerio, M.J.; Nichols, R.A. Dates from the molecular clock: How wrong can we be? Trends Ecol. Evol. 2007, 22, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.M.; Lajeunesse, M.J.; Rohr, J.R. A global synthesis of animal phenological responses to climate change. Nat. Clim. Chang. 2018, 8, 224–228. [Google Scholar] [CrossRef]
- Vaidya, M.M.; Kumar, P.; Singh, S.V. Circadian changes in heat storage and heat loss through sweating and panting in Karan Fries cattle during different seasons. Biol. Rhythm Res. 2011, 43, 137–146. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I. Genetic component of heat stress in dairy cattle, parameter estimation. J. Dairy Sci. 2000, 83, 2126–2130. [Google Scholar] [CrossRef]
- Jeelani, R.; Konwar, D.; Khan, A.; Kumar, D.; Chakraborty, D.; Brahma, B. Reassessment of temperature-humidity index for measuring heat stress in crossbred dairy cattle of a sub-tropical region. J. Therm. Biol. 2019, 82, 99–106. [Google Scholar] [CrossRef]
- Brode, P.; Fiala, D.; Blazejczyk, K.; Holmer, I.; Jendrizky, G.; Kampmann, B.; Tinz, B.; Havenith, G. Deriving the operational procedure for the universal thermal climatic index (UTCI). Int. J. Biomateorol. 2012, 56, 481–494. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I. Studies on genetics of heat tolerance in dairy cattle with reduced weather information via cluster analysis. J. Dairy Sci. 2002, 85, 1586–1589. [Google Scholar] [CrossRef]
- Bohmanova, J.; Misztal, I.; Cole, J.B. Temperature humidity indices as indicators of milk production losses due to heat stress. J. Dairy Sci. 2006, 90, 1947–1956. [Google Scholar] [CrossRef]
- Weber, J.A.; Park, S.G.; Luria, V.; Jeon, S.; Kim, H.M.; Jeon, Y.; Bhak, Y.; Jun, J.H.; Kim, S.W.; Hong, W.H.; et al. The whale shark genome reveals how genomic and physiological properties scale with body size. Proc. Natl. Acad. Sci. USA 2020, 117, 20662–20671. [Google Scholar] [CrossRef]
- Blache, D.; Maloney, S.K. New physiological measures of the biological cost of responding to challenges. In Advances in Sheep Welfare; Ferguson, D.M., Lee, C., Fisher, A., Eds.; Elsevier: London, UK, 2017; pp. 73–104. [Google Scholar]
- Gauly, M.; Ammer, S. Challenges for dairy cow production systems arising from climate changes. Animals 2020, 14, 196–203. [Google Scholar] [CrossRef]
- Yadav, B.; Singh, G.; Wankar, A. Acclimatization dynamics to extreme heat stress in crossbred cattle. Biol. Rhythm Res. 2021, 52, 524–534. [Google Scholar] [CrossRef]
- Sammad, A.; Wang, Y.J.; Umer, S.; Lirong, H.; Khan, I.; Khan, A.; Ahmad, B.; Wang, Y. Nutritional physiology and biochemistry of dairy cattle under the influence of heat stress: Consequences and opportunities. Animals 2020, 10, 793. [Google Scholar] [CrossRef]
- Archana, P.R.; Aleena, J.; Pragna, P.; Vidya, M.K.; Niyas, A.P.A.; Bagath, M.; Krishnan, G.; Manimaran, A.; Beena, V.; Kurien, E.K.; et al. Role of heat shock proteins in livestock adaptation to heat stress. J. Dairy Vet. Anim. Res. 2017, 5, 13–19. [Google Scholar]
- Murphy, K.T.; Petersen, A.C.; Goodman, C.; Gong, X.; Leppik, J.A.; Garnham, A.P.; Cameron-Smith, D.; Snow, R.J.; McKenna, M.J. Prolonged submaximal exercise induces isoform-specific Na+-K+-ATPase mRNA and protein responses in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, 414–424. [Google Scholar] [CrossRef][Green Version]
- Wyckelsma, V.L.; Perry, B.D.; Bangsbo, J.; McKenna, M.J. Inactivity and exercise training differentially regulate abundance of Na+-K+-ATPase in human skeletal muscle. J. Appl. Physiol. 2019, 127, 905–920. [Google Scholar] [CrossRef]
- Sahoo, S.S.; Mishra, C.; Rout, M.; Nayak, G.; Mohanty, S.T.; Panigrahy, K.K. Comparative in silico and protein-protein interaction network analysis of ATP1A1 gene. Gene Rep. 2016, 5, 134–139. [Google Scholar] [CrossRef]
- Signor, S.A.; Nuzhdin, S.V. The evolution of gene expression in cis and trans. Trends Genet. 2018, 34, 532–544. [Google Scholar] [CrossRef]
- Kaushik, R.; Goel, A.; Rout, P.K. Differential expression and characterization of ATP1A1 exon17 gene by high resolution melting analysis and RT-PCR in Indian goats. Mol. Biol. Rep. 2019, 46, 5273–5286. [Google Scholar] [CrossRef]
- Kaur, R.; Sharma, A.; Sodhi, M.; Swami, S.K.; Sharma, V.L.; Kumari, P.; Verma, P.; Mukesh, M. Sequence characterization of alpha 1 isoform (ATP1A1) of Na+/K+-ATPase gene and expression characteristics of its major isoforms across tissues of riverine buffaloes (Bubalus bubalis). Gene Rep. 2018, 10, 97–108. [Google Scholar] [CrossRef]
- Idris, M.; Uddin, J.; Sullivan, M.; McNeill, D.M.; Phillips, C.J. Non-invasive physiological indicators of heat stress in cattle. Animals 2021, 11, 71. [Google Scholar] [CrossRef]
- Moorjani, P.; Amorim, C.E.G.; Arndt, P.F.; Przeworski, M. Variation in the molecular clock of primates. Proc. Natl. Acad. Sci. USA 2016, 113, 10607–10612. [Google Scholar] [CrossRef]
- Loeza-Quintana, T.; Adamowicz, S.J. Iterative calibration: A novel approach for calibrating the molecular clock using complex geological events. J. Mol. Evol. 2018, 86, 118–137. [Google Scholar] [CrossRef]
- Di Marco, M.; Pacifici, M.; Maiorano, L.; Rondinini, C. Drivers of change in the realised climatic niche of terrestrial mammals. Ecography 2021, 44, 1–11. [Google Scholar] [CrossRef]
- Bulmer, M.; Wolfe, K.H.; Sharp, P.M. Synonymous nucleotide substitution rates in mammalian genes: Implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. USA 1991, 88, 5974–5978. [Google Scholar] [CrossRef]
- Mignone, F.; Gissi, C.; Liuni, S.; Pesole, G. Untranslated regions of mRNAs. Genome Biol. 2002, 3, 1–10. [Google Scholar] [CrossRef]
- Baker, E. mRNA stability and localization. In Lewin’s Genes X, 10th ed.; Lewin, B., Krebs, J.E., Goldstein, E.S., Kilpatrick, S.T., Eds.; Jones and Bartlett Learning: London, UK, 2011; pp. 618–639. [Google Scholar]
- Genuth, N.R.; Barna, M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Meyer, S.; Temme, C.; Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 2017, 51, 171–194. [Google Scholar] [CrossRef] [PubMed]
- Tomanek, L. Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs. J. Exp. Biol. 2010, 213, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Fuller, A.; Mitchell, D.; Maloney, S.K.; Hetem, R.S.; Fonsêca, V.F.; Meyer, L.C.; van de Ven, T.M.; Snelling, E.P. How dryland mammals will respond to climate change: The effects of body size, heat load and a lack of food and water. J. Exp. Biol. 2021, 224, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Renaudeau, D.; Collin, A.; Yahav, S.; De Basilio, V.; Gourdine, J.L.; Collier, R.J. Adaptation to hot climate and strategies to alle viate heat stress in livestock production. Animals 2012, 6, 707–728. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).