Simple Summary
The dead end (dnd) gene encodes an RNA-binding protein that plays a role for migration of primordial germ cells (PGCs) to the gonadal region during embryogenesis in vertebrates. Here, the starry flounder (Platichthys stellatus) dead end (psdnd) gene was characterized and its expression patterns were analyzed. Full-length psdnd mRNA was 1495 bp long, encoding 395 amino acids. psdnd was only expressed in gonadal tissues, we detected no psdnd expression in somatic cells. Furthermore, psdnd was strongly expressed during early embryogenesis. Our findings suggest that psdnd expression is gonad-specific and could therefore be used as a germ cell marker in starry flounder.
Abstract
dnd is a germline-specific maternal RNA expressed in various vertebrate classes, which encodes an RNA-binding protein that is essential for PGC migration. The purpose of this study is fundamental research about starry flounder dnd gene for germ cell marker development. In this study, we cloned and analyzed the expression levels of Platichthys stellatus dead end (psdnd) in various tissues and embryonic stages. The psdnd gene was isolated from starry flounder ovaries, cloned into a pGEM-t vector, and sequenced. Full-length of psdnd cDNA was 1495 bp long, encoding 395 amino acids. psdnd expression levels were investigated by real-time polymerase chain reaction (qRT-PCR) in various tissues and embryo developmental stages. psdnd transcripts were detected in the testes and ovaries, but not in somatic tissues. Embryonic psdnd expression levels were higher during early embryo development stages than during late embryogenesis; psdnd expression was highest at the 1 cell stage, then gradually decreased throughout the subsequent developmental stages. The spatial expression pattern was analyzed by whole-mount in situ hybridization (WISH). The psdnd transcripts migration pattern was similar with zebrafish (Danio rerio). Our results suggest that psdnd may function as a germ cell-specific marker.
1. Introduction
PGCs are the only cells that can transmit genetic material to the next generation. PGCs move to the gonadal location during early embryogenesis where they develop into gonads [1]. The proliferation of PGCs become the germline stem cells which can develop to mature gonads. Furthermore, migration of PGCs impacts on the fertility [2]. The cell fate and development of PGCs are determined by germ plasm mRNA derived from the maternal line [2]. Maternal germplasm mRNAs encode evolutionarily conserved proteins, including vasa, nanos C2HC-type zinc finger 3 (nanos3), dead-end (dnd), tudor-domain-containing protein 7 (tdrd7), and deleted in azzospermia-like (dazl) [2,3,4,5,6].
dnd is a germ plasm-specific marker in several vertebrate species that encodes an RNA-binding protein essential for PGC migration [7]. Uracil-rich sequences in the microRNA 430 bind to DND in zebrafish (Danio rerio). dnd prevents the degradation of germ plasm mRNAs targeted by mi430, thus maintaining germ cell development [8].
Germ cell-specific expression of dnd was first discovered in zebrafish after which similar expression patterns were found in various vertebrate species including mice (Mus musculus), frogs (Xenopus tropicalis), chickens (Gallus gallus), and humans (Homo sapiens) [8,9,10,11]. Thus, dnd is an evolutionarily conserved gene. In zebrafish and medaka (Oryzias latipes), germ cells are essential for female gonadal differentiation and the absence of PGCs induces sterility in males [12,13]. In addition, the removal of PGCs from pond loach (Misgurnus anguillicaudatus) and goldfish (Carassius auratus) induces sterility both males and females [14,15]. Abnormal PGC localization was induced in zebrafish by knocking down dnd expression [12]. In frogs, PGCs were lost when dnd was knocked down using morpholino oligonucleotide [16]. These studies indicate that dnd is essential for germ cell development during embryogenesis.
Most farmed fish reach gonadal maturation before they are released to the market, and gonadal maturation is an important factor that determines the marketability of fish [17]. However, infertile fish exhibit improved growth rates, meat quality, and disease resistance; these effects are achieved by minimizing the energy required for gonadal development [18,19,20,21,22]. In addition, sterilization is an effective method to reduce ecological genetic contamination by protecting natural species from escaped farmed fish, thus maintaining genetic diversity [17].
The development of sterilization strategies is an important goal for researchers. To achieve this, genetic modification techniques such as chromosome manipulation, transgene expression, morpholino oligonucleotide treatment, and CRISPR/Cas9 are increasingly undergoing development and application in the aquaculture industry. dnd knockdowns can induce sterility in organisms by blocking PGC migration to the gonad developmental region. Genetic engineering techniques have been applied to knock-down dnd in channel catfish (Ictalurus punctatus) [23]. Furthermore, salmon were successfully sterilized by targeting dnd with CRISPR/Cas9 [24]. Infertility was induced in sterlet (Acipenser ruthenus) by knocking down dnd with an antisense morpholino oligonucleotide [25].
To support novel technologies for producing infertile eggs in starry flounder, we conducted fundamental research including identification of psdnd and expression pattern during embryogenesis. Improving our understanding of the mechanisms driving gonad development, such as dnd expression, would enable the development of new sex maturation control technologies.
2. Materials and Methods
2.1. Animals and Sampling
3 females and 3 males which fully matured starry flounders were used in this experiment from Marine seed fish farm (Yeosu, Korea). These fishes were raised in a 15 ton tank controlling the water temperature. The mean total length and weight of females was 33.8 ± 2.8 cm and 825.7 ± 186.5 g. The mean total length and weight of males was were 32.6 ± 0.9 cm and 504.6 ± 87.4 g. Starry flounders were kept on 16:8 light:dark cycle. Exogenous salmon gonadotropin-releasing hormone analog (sGnRHa) (Ovaplant, Syndel, Ferndale, WA, USA) pellets were inserted into the dorsal muscles at a concentration of 50 µg/kg. Various tissues including brain, gill, heart, kidney, eye, stomach, gut, spleen, liver, muscle, testis, and ovary were collected from fully matured female and male starry flounders. Fertilized eggs were cultured at 10 ± 1 °C after artificial insemination. Embryo samples including unfertilized egg, 1 cell, 2 cell, 4 cell, 8 cell, 16 cell, morula, blastula, early gastrula, late gastrula, somite, and hatching larva were collected for analysis. Tissues and embryos were stored in liquid nitrogen prior to RNA extraction.
2.2. Total RNA Extraction and cDNA Synthesis
Total RNA was extracted followed by the manufacturer’s instructions. Total RNA was extracted from 100 mg of each tissue (brain, gill, heart, kidney, eye, stomach, gut, spleen, liver, muscle, testis, and ovary) using TRIzol® (Invitrogen, Waltham, MA, USA). In addition, total RNA was extracted from 100 mg of embryo tissue at 12 distinct developmental stages using TRIzol® reagent. 200 μL of chloroform were added and reacted at −20 °C for 5 min. Centrifugation at 12,000 rpm, 4 °C for 15 min. The supernatant was mixed with the same amount of isopropanol and reacted for 10 min. Centrifuge at 12,000 rpm, 4 °C for 10 min. The supernatant was removed and 1 mL of 70% ethanol was added for washing the pellet. The ethanol was removed completely by centrifuging at 12,000 rpm, 4 °C for 5 min. 100 μL of diethylpyrocarbonate (DEPC) water was added to the pellet and resuspended. DNase I (Qiagen, Hilden, Germany) was treated to prevent DNA contamination. cDNA was synthesized from purified total RNA using a Maxima First Strand cDNA Synthesis Kit (Thermo, Waltham, MA, USA). Two microliters of Maxima Enzyme Mix, 4 μL of 5 × reaction mix, and 14 μL of RNA were mixed for real-time PCR. The cDNA synthesis conditions were 25 °C for 10 min, 65 °C for 30 min, and 85 °C for 5 min. The synthesized cDNA was quantified using a NANODROP ONEC (Thermo, Waltham, MA, USA) and stored in a deep freezer (−80 °C).
2.3. PCR Amplification of cDNA Fragment
Primers were designed based on the dnd mRNA sequence of olive flounder to detect and amplify psdnd. The olive flounder dnd sequence (Accession No. KP224455.1) was obtained from the National Center for Biotechnology database (Table 1).

Table 1.
Primer and probe sequences for psdnd cloning and qRT-PCR.
The PCR cycling conditions were as follows: pre-denaturation at 94 °C for 5 min; 30 cycles of 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 2 min; then a final extension at 72 °C for 10 min. The PCR products were analyzed by electrophoresis with a 1% agarose gel. The DNA was gel extracted and sequenced by Macrogen Inc. (Seoul, Korea).
2.4. 5′ and 3′ Rapid Amplification of cDNA Ends (RACE) PCR
5′ and 3′ gene-specific primers (GSPs) were designed based on the partial fragment of starry flounder dnd to obtain the full transcript (Table 1). 5′ RACE PCR was conducted using a 5′ RACE system for Rapid Amplification of cDNA Ends kit version 2.0 (Invitrogen, Waltham, MA, USA), in accordance with the manufacturer’s instructions.
3’ cDNA was synthesized using a SMART cDNA synthesis kit (Clontech, MountainView, CA, USA). Total RNA was isolated from starry flounder ovarian tissues using TRIzol® reagent. Single-step PCR and semi-nested PCR were performed using the synthesized 3’ cDNA. One microliter of 3’ GSP1 primer, 1 µL of universal primer mix, and 2 μL of 3’ cDNA were added to the reaction mixture containing 0.25 μL of Advantage Taq polymerase (Takara, Nojihigashi, Japan), 1.5 μL of 10 × buffer, 1 μL of dNTPs, and sterile water to a total volume of 10 μL. The single-step PCR cycling conditions were as follows: pre-denaturation at 94 °C for 2 min 30 s; 35 cycles of 94 °C for 30 s, annealing at 58 °C for 30 s, and amplification at 72 °C for 2 min; then a final extension at 72 °C for 5 min. The semi-nested PCR was conducted with 1 μL of nested universal primer, 1 μL of 3’ GSP2, 1 μL of the single-step PCR product, 17 μL of 1.1 × master mix, and sterile water to a total volume of 20 μL. The semi-nested PCR was performed using thermocycling conditions identical to the conditions used for single-step PCR. PCR products were an-alyzed by means of electrophoresis and sequencing, performed by Macrogen Inc. (Seoul, Korea).
2.5. Cloning
Primers were designed based on the 1525 bp psdnd cDNA sequence obtained via 5’ and 3’ RACE PCR. One microliter of ovary cDNA was amplified with AccuPower® PCR premix (Bioneer, Deajeon, Korea) and 10 pM of each primer (sense and antisense). The PCR cycling parameters were as follows: initial denaturing at 94 °C for 5 min; 35 cycles of denaturing at 94 °C for 30 s, annealing at 53 °C for 30 s, and elongation at 72 °C for 2 min; then a final elongation at 72 °C for 10 min. PCR products were analyzed on a 1% agarose gel and DNA within a size range of 1400–1600 bp was extracted using a Gel extraction kit (Bioneer, Korea). cDNA (30 ng/μL) was ligated into the pGEM-T vector system I (Promega, Madison, WI, USA), in accordance with the manufacturer’s instructions. Ten microliters of ligation mixture were used to transform 50 μL of Escherichia coli DH5α Competent Cells (Takara, Nojihigashi, Japan), and the cells were incubated at 37 °C for recovery. The transformation culture was then incubated for 16 h at 37 °C on MacConkey agar supplemented with ampicillin for colony selection. Plasmid DNA was extracted using an Accuprep® plasmid Mini extraction kit (Bioneer, Korea). The extracted plasmid DNA was sequenced by Macrogen Inc. (Seoul, Korea).
2.6. Phylogenetic Analysis of psdnd
Molecular phylogenetic analysis was performed to compare the dnd sequences of different vertebrate species. The full dnd gene sequences of various species were retrieved from the NCBI database. The GenBank accession numbers of the sequences examined were as follows: atlantic salmon (Salmo salar), JN712911.1; channel catfish (Ictalurus punctatus), XM_017484732.1; chicken (Gallus gallus), XM_015293536.2; common carp (Cyprinus carpio), XM_019103334.1; atlantic halibut (Hippoglossus hippoglossus), XM_034598806.1; human (Homo sapiens), NM_194249.3; pond loach (Misgurnus anguillicaudatus), MH283870.1; mouse (Mus musculus), NM_173383.2; olive flounder (Paralichthys olivaceus), KP224455.1; pacific bluefin tuna (Thunnus orientalis), KF128758.1; rainbow trout (Oncorhynchus mykiss), MK887177.1; turbot (Scophthalmus maximus), KC460339.1; western clawed frog (Xenopus tropicalis), NM_001044434.1 and zebrafish (Danio rerio), AY225448.1.
A phylogenetic tree was constructed with the Mega X version 10.1 software using the maximum-likelihood method with a bootstrap analysis of 1000 replicates. The percentage identities between the starry flounder dnd sequence and the homologues genes from other species were analyzed with CLUSTALW.
2.7. qRT-PCR
qRT-PCR was performed using a Pikoreal 96 RealTime PCR System (Thermo, Waltham, MA, USA) to examine the psdnd expression profiles in various tissues and stages of embryo development. The concentrations of cDNA from eight distinct tissues and 12 developmental stages were adjusted to 50 ng/μL and 500 ng/μL respectively. The primers used for qRT-PCR are listed in Table 1. dnd transcripts were detected using SYBR Green PCR Mastermix (Thermo, Waltham, MA, USA), mixed with 1 μL of each primer (10 pM), 1 μL of cDNA, and 7 μL of sterile water. The qRT-PCR cycling conditions were as follows: pre-incubation at 50 °C for 2 min; pre-denaturation at 95 °C for 7 min; and then 40 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 20 s. Expression levels of the housekeeping gene glyceraldehyde3-phosphate dehydrogenase were used to calculate the relative expression levels of psdnd. psdnd expression levels in the spleen and fertilized eggs were used to calculate Ct values. Relative expression levels were determined by calculating 2−ΔΔCt using mean Ct values. The qRT-PCR primer sequences are shown in Table 1.
2.8. Whole Mount In Situ Hybridization (WISH)
Digoxigenin-labeled antisense probes were synthesized from linearized plasmids containing the dnd sequence using a DIG RNA Labeling kit (Roche, Basel, Switzerland). The primers used for WISH are shown in Table 1. Embryo samples were fixed in 4% paraformaldehyde with phosphate-buffered saline. The embryo samples were dehydrated using 50% and 30% methanol, and the pigments were removed from the embryo samples using 6% hydrogen peroxide and 0.5% potassium hydroxide. Then, proteinase K solution (10 μg/mL) was applied to permeabilize embryo membranes, and the embryo samples were fixed again in 4% paraformaldehyde. Samples were incubated in hybridization solution at 65 °C for 3 h; 1 μg/mL antisense RNA probe was then added to the hybridization solution and incubated at 85 °C for 3 minutes. A longer hybridization reaction was also conducted at 65 °C for 12 h. The dnd transcripts were visualized using purple alkaline phosphatase substrate.
2.9. Statistical Analysis
The qRT-PCR results were expressed as mean ± SD. Statistical analyses were performed using SPSS Statistic version 26.0 (SPSS Inc., Chicago, IL, USA). One-way analysis of variance and Tukey’s test were used to determine significance (p < 0.05).
3. Results
3.1. Identification of P. stellatus cDNA Sequence
The full-length psdnd mRNA was 1495 bp long, containing an 1185 bp coding sequence encoding 395 amino acids (Figure 1). psdnd cDNA had five exon regions; the length of exons 1 to 5 were 151 bp, 111 bp, 327 bp, 133 bp, and 536 bp respectively. The length of the 5′-untranslated region was 73 bp. The 3′-untranslated region was 239 bp long, including the complete 3′ end with a poly-(A)-tail. psdnd genomic DNA had 4 introns with lengths of 1240 bp, 1001 bp, 751 bp, and 1509 bp (Figure 2).

Figure 1.
Nucleotide and amino acid sequence of psdnd. The first methionine amino acid indicates the start codon. Strock indicates the stop codon. Amino acid sequence was analyzed by clustal omega website available at https://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 15 March 2020).
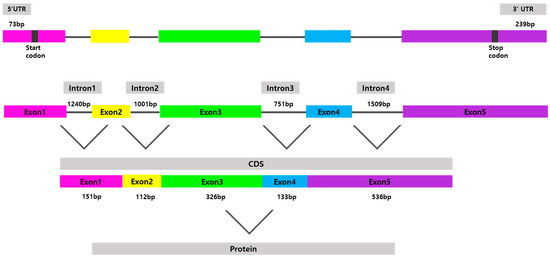
Figure 2.
Physical map of cloned psdnd. The psdnd open reading frame (ORF) contains 5 exons and 4 introns. Revealed 73 bp of 5′UTR and 239 bp of 3′UTR. Each exons are shown in different color boxes. Introns expressed thin grey lines.
3.2. Multiple Sequence Alignments
A multiple alignments comparing the amino acid sequence deduced from psdnd with the dnd sequences of other species is shown in Figure 3. The range of sequence identities of the dnd amino acid sequences was 31–85% (Table 2). Of the alignment, data suggested that several regions of the dnd protein were conserved between species. The amino acid sequence determined using the psdnd cDNA sequence possessed three conserved motifs including two RNA-recognition motifs (RRMs) and one double-stranded RNA-binding motif (DSRM). The RRMs and DSRM were shared among dnd homologues.

Figure 3.
Multiple alignments of psdnd with dnd homologues from various vertebrate species. Black text on white: non-similar residues. Blue text on cyan: consensus residue, derived from a block of similar residues at a specific position. Black text on green: consensus residue, derived from an occurrence of ≥50% of a single residue at a specific position. Red on yellow: consensus residue, derived from a completely conserved residue at a specific position. Green text on white: residue similar to consensus residue at a specific position. Red boxes indicate conserved motifs including RRM1, RRM2 and, DSRM. Multiple alignment analyzed by align X program. Conserved domain was analyzed at NCBI conserved domain site.

Table 2.
Sequence identities comparing psdnd to homologues in other species.
3.3. Phylogenetic Tree and Identity
The deduced psdnd amino acid sequence was aligned with related protein sequences from various vertebrate species (Table 2). According to the neighbor-joining tree shown in Figure 4, psdnd and atlantic halibut dnd formed a clade supported by a bootstrap value of 100%. psdnd exhibited identities of 85%, 77%, 62%, 61%, 50%, 49%, 39%, 39%, 37%, 35%, 33%, 32%, 32%, and 31% with dnd homologues in atlantic halibut, olive flounder, turbot, pacific bluefin tuna, rainbow trout, Atlantic salmon, pond loach, channel catfish, common carp, zebrafish, human, mouse, chicken, and western clawed frog respectively (Figure 4).
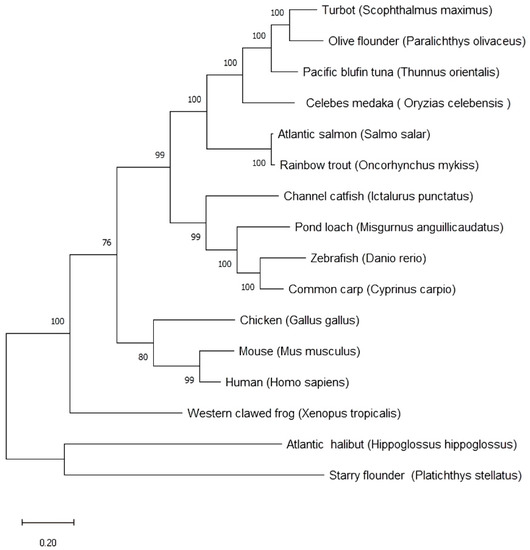
Figure 4.
Phylogenetic tree and sequences identities comparing psdnd with dnd from various vertebrate species. Phylogenetic tree of psdnd established by MEGAX program with maximum likelihood method. The numbers under the nodes indicate bootstrap percentage values for 1000 replicates. Scale bar indicates number of substitution changes per site. Bootstrap values indicate branch.
The highest protein sequence identity is 85% of Atlantic halibut. As followed DND protein identity ranked high 77% of olive flounder and 62% of turbot. The lowest protein sequence identity is 31% of western clawed frog.
3.4. qRT-PCR
psdnd transcripts were detected in both male and female gonads by means of qRT-PCR. However, psdnd expression was significantly greater in the ovaries than in the testes. No psdnd expression was detected in the other tissues examined (Figure 5a).
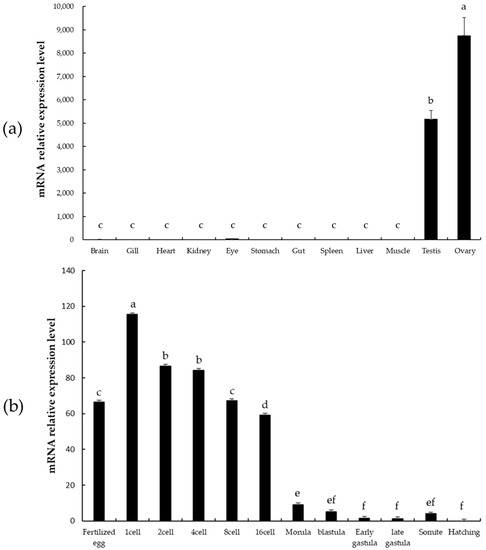
Figure 5.
psdnd transcript expression levels in various tissues and embryo stages measured by qRT-PCR. (a) Tissue-specific expression of psdnd (p < 0.05). (b) Embryonic expression of psdnd. Different letters indicate statistically significant difference (p < 0.05).
psdnd mRNA was expressed during all stages of embryonic development and expression was maintained until hatching (Figure 5b). Strong expression was detected from the fertilized egg stage until the 16 cell stage. After fertilization, psdnd transcript expression increased and peaked at the 1 cell stage. psdnd expression decreased slightly until 16 cell stage, then decreased dramatically from the 16 cell stage to the hatching stage.
3.5. Whole-Mount In Situ Hybridization
Two spots of dnd positive cells were detected at the edges of the first furrows during the 2 cell stage (Figure 6a). psdnd was expressed in four spots at the ends of the second furrows from the 4 cell to the morula stage (Figure 6b–d). At the blastula stage, multiple sister cells were generated close to the psdnd positive-cells (Figure 6e). During the early gastrula stage, psdnd expression was detected at the blastoderm margins (Figure 6f). At the late gastrula stage, few psdnd transcripts were dispersed however they were on move at the trunk mesoderm (Figure 6g). The psdnd positive signals then moved toward the dorsal mesentery, where the gonads would later develop; psdnd expression continued in this area until the hatching stage (Figure 6h,i).
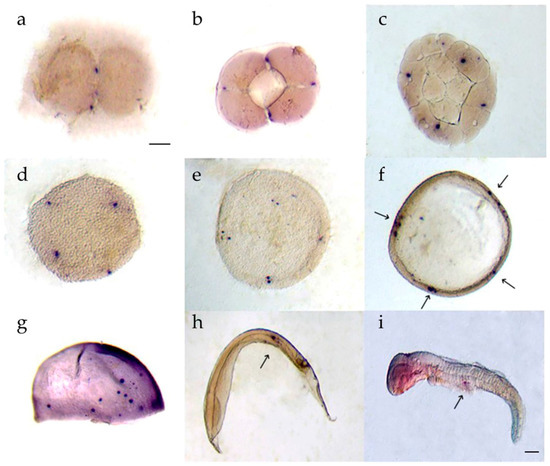
Figure 6.
psdnd transcript localization during embryogenesis. Embryos were hybridized with antisense psdnd, stained by purple AP. (a) 2 cell stage: psdnd transcripts presented at first cleavage site; (b) 4 cell stage: psdnd positive signals revealed the spots where edges of second cleavage; (c) 32 cell stage; (d) morula stage: four dnd positive signals maintained until morula stage; (e) blastula stage: sister cells developed next to origin cells; (f) early gastrula stage; dnd transcripts increased and clustered at four spots of germ rings; (g) late gastrula stage; psdnd positive signals were move on to body axes; (h) early somite stage; (i) hatching larva: psdnd transcripts were aggregated and move on to future gonad location. Arrows indicate psdnd transcript expression. Scale bar: 100 μm.
4. Discussion
In this study, the psdnd protein sequence contains two RRM motif and one DSRM motif at its C-terminus, which are characteristic of DND protein [1]. The interspecific similarities in the DND amino acid sequences suggest that they have similar structures and functions, and that DND is well conserved. The first RRM motif, RRM1, is the main binding site for DND1 protein that directly binds to nanos1. The second RRM motif, RRM2, plays a role in promoting nanos1 translation and germline survival in xenopus [26]. DSRM is not essential for RNA binding, although it increases the target specificity of the RRM regions in DND1 in human [27]. Zebrafish dnd has a binding site to bind miR-430 for preventing degradation from miRNA [8]. However, the structure and function of which has not been evaluated in starry flounder. Further study is needed to understand the relationship between psdnd and miR-430 in starry flounder.
The psdnd protein sequence exhibited strong homologues with DND proteins of other species, especially other flatfish species such as Atlantic halibut (85%), olive flounder (77%), and turbot (62%). The phylogenetic tree and identity results indicated that dnd in starry flounder and Atlantic halibut have the highest evolutionary closeness in our researched target species.
The transcript expression of psdnd was analyzed in various tissues by means of qRT-PCR. psdnd expression was only detected in the ovaries and testes, suggesting that psdnd can be used as a germ cell marker in starry flounder. In some species, such as mouse and xenopus, dnd expression is sex-dependent [13,28]. However, in fish species, dnd is expressed in both sexes, as observed in this study [7,13,18,29,30]. The expression in both ovary and testis means psdnd has specific roles for oogenesis and spermatogenesis during sexual maturation in starry flounder.
The expression levels of psdnd at various embryo developmental stages were also investigated by qRT-PCR. We found that psdnd was highly expressed during the early developmental stages, from the 1 cell to the early gastrula stage. Higher expression patterns during early embryogenesis also revealed in olive flounder and turbot [7,28]. This suggests that embryonic psdnd expression is of maternal origin, similar with the germplasm marker, vasa. psdnd was also detected at the zygote stage. After the gastrula stage, psdnd transcripts began to decrease more dramatically. Maternal genes are often highly expressed until the zygotic genome is activated. Depending on the genes or species, continuously sex-pressed gene can interact with the zygotic genome [31]. The pattern of psdnd expression pattern may be related to the expression of the microRNA 430. MicroRNAs are small molecules that regulate the expression of target mRNAs by degradation. In zebrafish, miR-430 targets nanos and tdrd7, which are essential for PGC development. DND1 prevents miR-430 from binding to its target mRNAs. After zygotic genome activation at the gastrula stage, miR-430 and DND1 expression levels decrease [8,32,33]. In this study, we found that psdnd was strongly expressed before the blastula stage, suggesting that psdnd was of maternal origin.
The successful migration of PGCs to the gonads is essential for PGC development and survival. PGCs require maternally supplied germplasm and mRNAs for their specification and maintenance. Our WISH results revealed that the psdnd expression pattern was similar with the PGC localization pattern, suggesting that psdnd is a germplasm involved in PGC migration. Similar quantitative and spatial expression patterns occur for dnd in other vertebrate species, including olive flounder, turbot, and rare minnow [7,30,31].
This finding of this study suggests that psdnd could be used as a germ cell marker in starry flounder; moreover, dnd could be a potential target for starry flounder sterilization.
5. Conclusions
In this study, we identified and cloned full-length psdnd, which was 1495 bp long and encoded 395 amino acids. psdnd was specifically expressed in starry flounder ovaries and testes. Moreover, psdnd was highly expressed during the early embryonic developmental stages, from fertilized egg to the 16 cell stage, after which expression significantly decreased. Our results suggest that psdnd is maternally derived and specifically expressed in germ cells, similar with dnd homologs in organisms.
Author Contributions
Conceptualization, J.-H.Y.; methodology, Y.-S.C. and J.-H.Y.; software, J.-H.Y., Y.-S.C.; formal analysis, J.-H.Y., Y.-S.C., H.-B.L. and J.-Y.P.; data curation, J.-H.Y.; writing—original draft preparation, J.-H.Y.; supervision, H.-K.L.; project administration, H.-K.L.; funding acquisition, H.-K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Korean Institute of Marine Science & Technology Promotion, Republic of Korea, with Assignment No. 20180373. This research was conducted as part of the project entitled “Establishing a foundation for the year-round production of flatfish eggs and improving productivity”.
Institutional Review Board Statement
The study was approved by the Institutional Animal Care and Use Committee of Mokpo National University No. 1183 (17 December 2013).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article.
Acknowledgments
The authors would like to thank Jum Sook Chung, Ten-Tsao Wong, and Jun Hyung Ryu, University of Maryland Baltimore County for methodological support and allowing us to work on University of Maryland. We thank Il Young Lee for support the resources.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baloch, A.R.; Franěk, R.; Saito, T.; Pšenička, M. Dead-end (dnd) protein in fish—A review. Fish. Physiol. Biochem. 2019, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Marlow, F. Primordial germ cell specification and migration. F1000Research 2015, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beer, R.L.; Draper, B.W. nanos3 maintains germline stem cells and expression of the conserved germline stem cell gene nanos2 in the zebrafish ovary. Dev. Biol. 2013, 374, 308–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komiya, T.; Itoh, K.; Ikenishi, K. Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev. Biol. 1994, 162, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Houwing, S.; Kaaij, L.J. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011, 30, 3298–3308. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.A.; Fulton, N.; Cowan, G. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev. Biol. 2007, 7, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, Q.; Xiao, Y.; Yang, Y.; Wang, Y.; Song, Z.; You, F.; An, H.; Xiao, Z.; Xu, S. The dnd RNA identifies germ cell origin and migration in olive flounder (Paralichthys olivaceus). BioMed Res. Int. 2015, 2015, 428591. [Google Scholar]
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Oude Vrielink, J.A.F.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Ørom, U.A.; et al. RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, C.; Aggarwal, S.; Zhu, R. The mouse dead-end gene isoform α is necessary for germ cell and embryonic viability. Biochem. Biophys. Res. Commun. 2007, 355, 194–199. [Google Scholar] [CrossRef] [Green Version]
- Shinomiya, A.; Tanaka, M.; Kobayashi, T.; Nagahama, Y.; Hamaguchi, S. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev. Growth Differ. 2000, 42, 317–326. [Google Scholar] [CrossRef]
- Aramaki, S.; Sato, F.; Kato, T.; Soh, T.; Kato, Y.; Hattori, M.A. Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res. 2007, 330, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, G.; Stebler, J.; Slanchev, K.; Dumstrei, K.; Wise, C.; Lovell-Badge, R.; Thisse, C.; Thisse, B.; Raz, E. Dead end, a Novel Vertebrate Germ Plasm Component, Is Required for Zebrafish Primordial Germ Cell Migration and Survival. Curr. Biol. 2003, 13, 1429–1434. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Hong, N.; Xu, H.; Li, M.; Yan, Y.; Purwanti, Y.; Yi, M.; Li, Z.; Wang, L.; Hong, Y. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr. Patterns 2009, 9, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Nishimura, T.; Goto-Kazeto, R.; Kawakami, Y.; Yamaha, E.; Arai, K. Sexual dimorphism of gonadal structure and gene expression in germ cell-deficient loach, a teleost fish. Proc. Natl. Acad. Sci. USA 2010, 107, 17211–17216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, R.; Saito, T.; Takeda, T.; Fujimoto, T.; Takagi, M.; Arai, K.; Yamaha, E. Germ cells are not the primary factor for sexual fate determination in goldfish. Dev. Biol. 2012, 370, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Horvay, K.; Claußen, M.; Katzer, M.; Landgrebe, J.; Pieler, T. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 2006, 291, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wong, T.T.; Zohar, Y. Production of reproductively sterile fish by a non-transgenic gene silencing technology. Sci. Rep. 2015, 5, 15822. [Google Scholar] [CrossRef] [Green Version]
- Hallerman, E.M.; Kapuscinski, A.R. Incorporating risk assessment and risk management into public policies on genetically modified finfish and shellfish. Aquaculture 1995, 137, 9–17. [Google Scholar] [CrossRef]
- Bartley, D.M.; Rana, K.; Immink, A.J. The use of inter-specific hybrids in aquaculture and fisheries. Rev. Fish. Biol. Fish. 2000, 10, 325–337. [Google Scholar] [CrossRef]
- Rahman, M.A.; Uehara, T.; Lawrence, J.M. Growth and heterosis of hybrids of two closely related species of Pacific sea urchins (Genus Echinometra) in Okinawa. Aquaculture 2005, 245, 121–133. [Google Scholar] [CrossRef]
- Kalsoom, U.; Salim, M.; Shahzadi, T.; Barlas, A. Growth performance and feed conversion ratio (FCR) in hybrid fish (Catla catle x Labeo rohita) fed on feed bran, rice broken and blood meal. Pak. Vet. J. 2009, 29, 55–58. [Google Scholar]
- Exadactylos, A.; Arvanitoyannis, I.S. Aquaculture Biotechnology for enhanced fish production for food consumption. In Microbial Biotechnology in Agriculture and Aquaculture, 2nd ed.; Ray, R.C., Ed.; Science Publishers Inc.: Enfield, NH, USA, 2006; pp. 453–510. [Google Scholar]
- Li, H.; Su, B.; Qin, G.; Ye, Z.; Elaswad, A.; Alsaqufi, A.; Perera, D.A.; Qin, Z.; Odin, R.; Vo, K. Repressible transgenic sterilization in channel catfish, Ictalurus punctatus, by knockdown of primordial germ cell genes with copper-sensitive constructs. Mar. Biotechnol. 2018, 20, 324–342. [Google Scholar] [CrossRef]
- Wargelius, A.; Leininger, S.; Skaftnesmo, K.O.; Kleppe, L.; Andersson, E.; Taranger, G.L.; Schulz, R.W.; Edvardsen, R.B. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 2016, 6, 21284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linhartová, Z.; Saito, T.; Kašpar, V.; Rodina, M.; Prášková, E.; Hagihara, S.; Pšenička, M. Sterilization of sterlet Acipenser ruthenus by using knockdown agent, antisense morpholino oligonucleotide, against dead end gene. Theriogenology 2015, 84, 1246–1255. [Google Scholar] [CrossRef] [Green Version]
- Aguero, T.; Jin, Z.; Owens, D.; Malhotra, A.; Newman, K.; Yang, J.; King, M.L. Combined functions of two RRMs in Dead-end1 mimic helicase activity to promote nanos1 translation in the germline. Mol. Reprod. Dev. 2018, 85, 896–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duszczyk, M.M.; Wischnewski, H.; Kazeeva, T.; Loughlin, F.E.; von Schroetter, C.; Pradère, M.U.; Ciaudo, C.; Allain, F.H.T. The solution structure of Dead End bound to AU-rich RNA reveals an unprecedented mode of tandem RRM-RNA recognition required for mRNA repression. BioRxiv 2019, 572156. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Collodi, P. Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development. FASEB J. 2010, 24, 2641–2650. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.; Zhao, C.Y.; Xu, S.H.; Ma, D.Y.; Xiao, Z.Z.; Xiao, Y.S.; Xu, C.A.; Liu, Q.H.; Li, J. Germline-specific and sexually dimorphic expression of a dead end gene homologue in turbot (Scophthalmus maximus). Theriogenology 2013, 80, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Feng, G.; Chang, P.; Zhang, X.; Zhou, Q.; Zhong, X.; Qi, C.; Xie, S.; Zhao, H. Germ cell-specific expression of dead end (dnd) in rare minnow (Gobiocypris rarus). Fish. Physiol. Biochem. 2015, 41, 561–571. [Google Scholar] [CrossRef]
- Omura, C.S.; Lott, S.E. The conserved regulatory basis of mRNA contributions to the early Drosophila embryo differs between the maternal and zygotic genomes. PLoS Genet. 2020, 16, e1008645. [Google Scholar] [CrossRef] [Green Version]
- Mishima, Y.; Giraldez, A.J.; Takeda, Y.; Fujiwara, T.; Sakamoto, H.; Schier, A.F.; Inoue, K. Differential Regulation of Germline mRNAs in Soma and Germ Cells by Zebrafish miR-430. Curr. Biol. 2006, 16, 2135–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).