A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Fish and Tissue Collection
2.3. Sequence Analysis and Alignment
2.4. cDNA Synthesis and Quantitative Real-Time PCR (qPCR) Analysis
2.5. Cellular Localization of Cs-rchy1 mRNA in Gonads
2.6. siRNA-Mediated Interference of Cs-rchy1 in Gonadal Cell Lines
3. Results
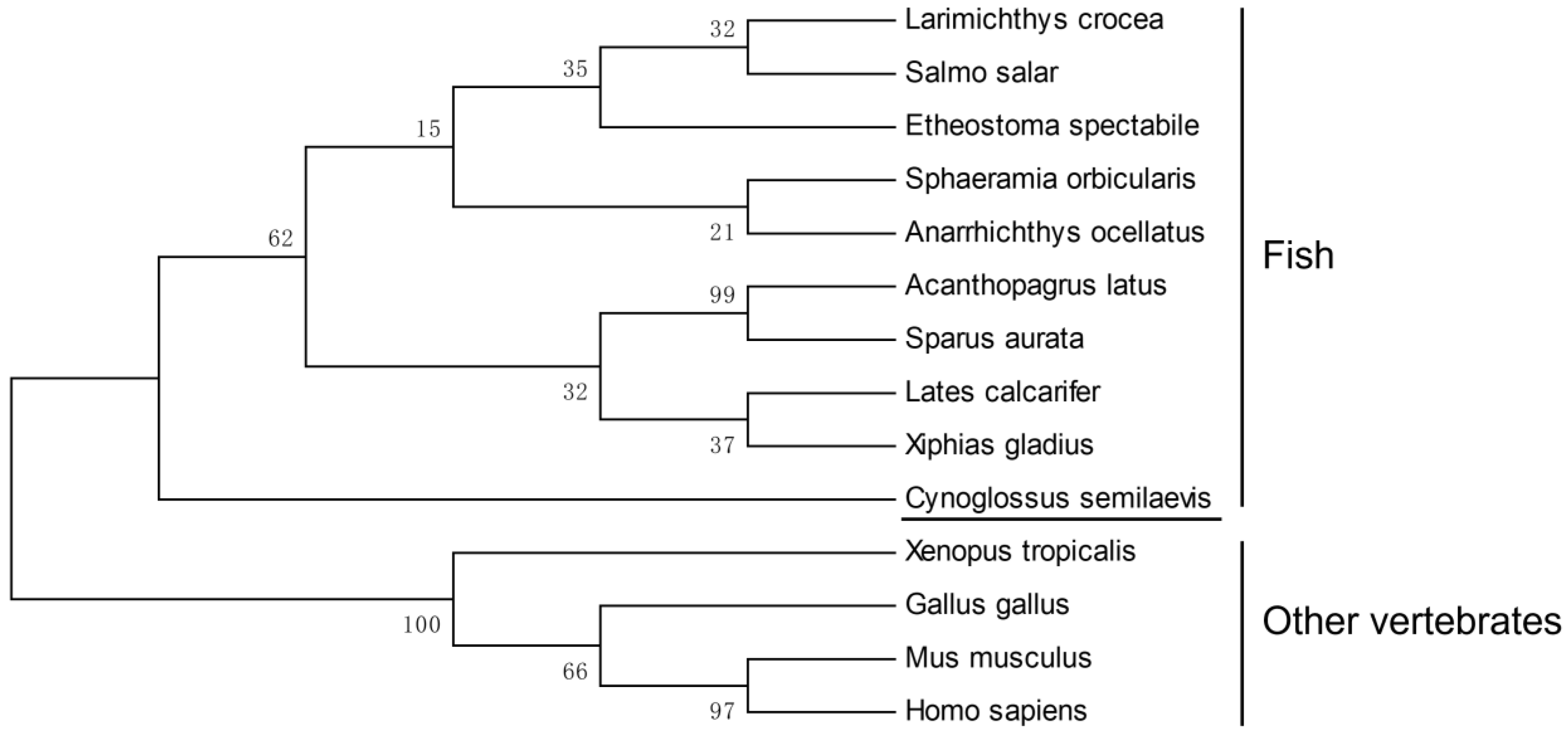
3.1. Cloning and Characteristics of CS-rchy1
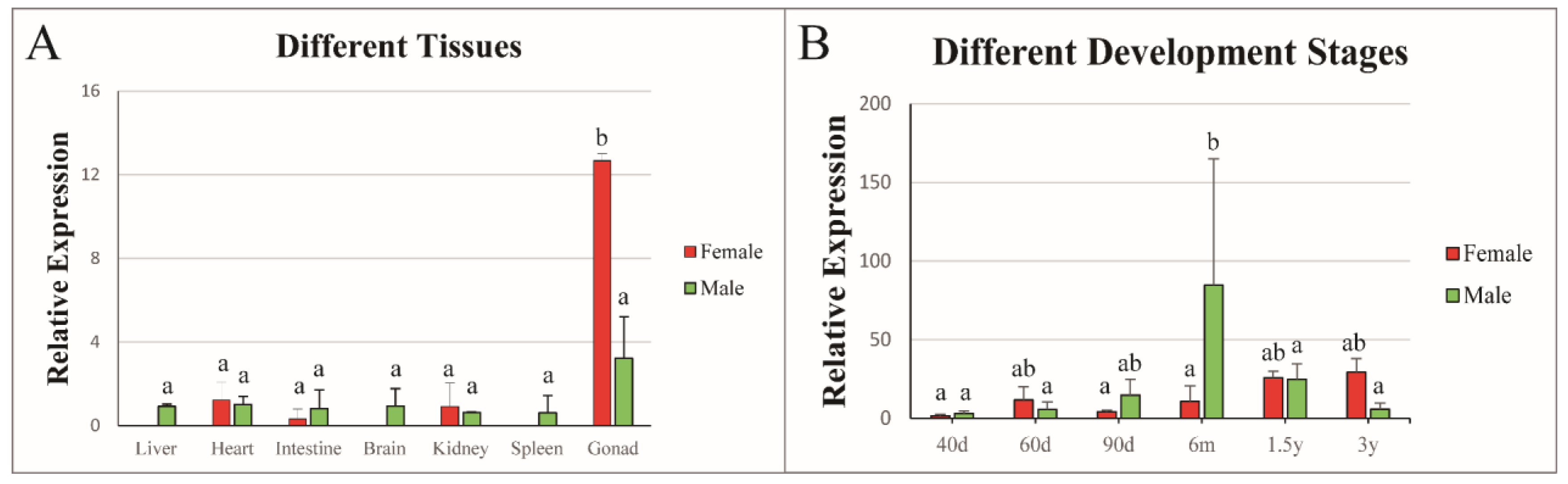
3.2. Tissue Expression Patterns of Cs-rchy1
3.3. Expression Profile of Cs-rchy1 at Different Developmental Stages of Gonads
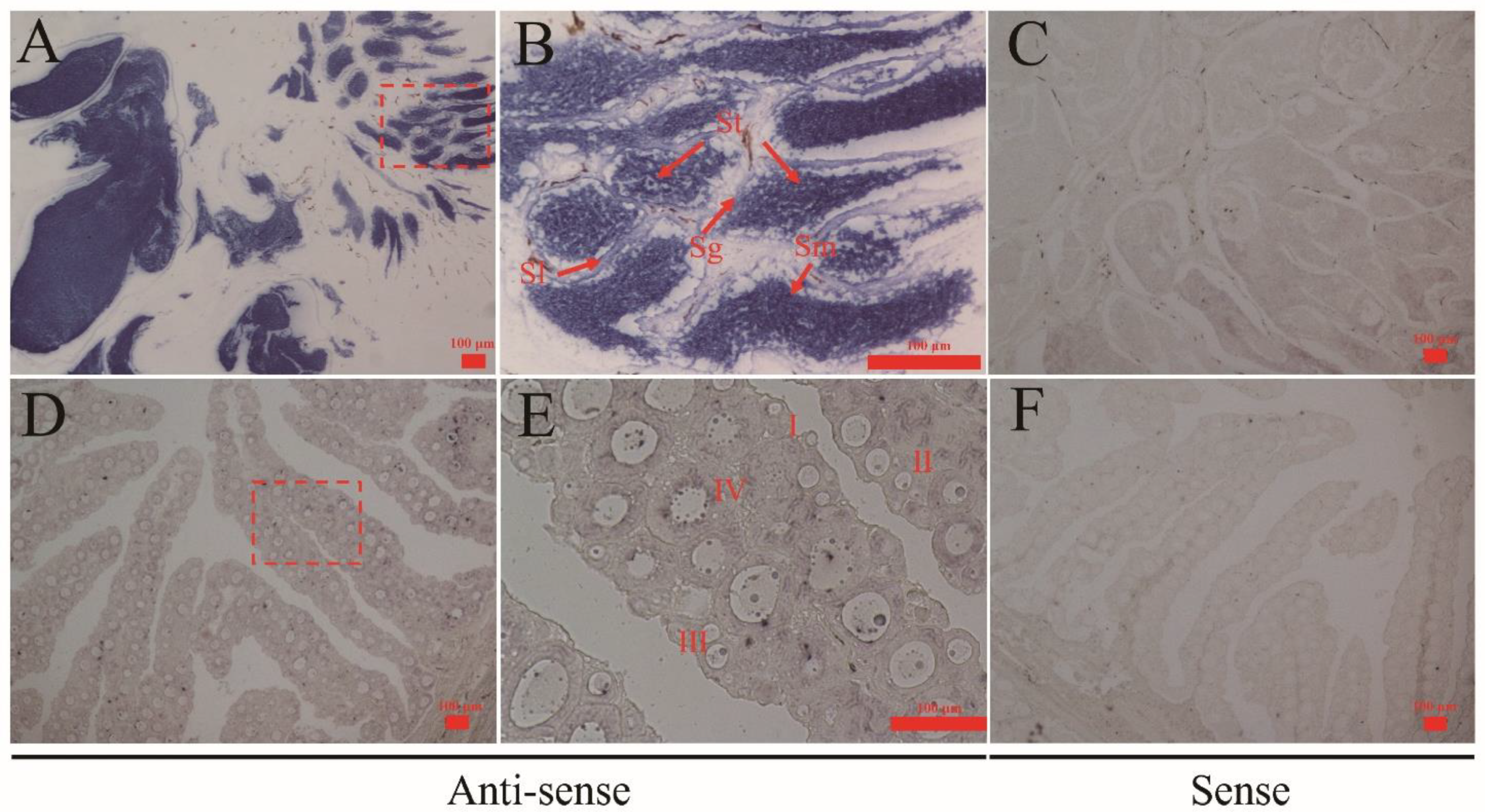
3.4. Localization of Cs-rchy1 mRNA in Gonads
3.5. In Vitro RNAi-Mediated Cs-rchy1 Knockdown and Its Influence on the Expression of Sex-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nat. Cell Biol. 1995, 373, 81–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.H.; Fang, W.; Hao, C.; Wei, S.; Zhang, X.S.; Wang, T. Transcript Profile Analyses of Maize Silks Reveal Effective Activation of Genes Involved in Microtubule-Based Movement, Ubiquitin-Dependent Protein Degradation, and Transport in the Pollination Process. PLoS ONE 2013, 8, e53545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtake, F.; Baba, A.; Takada, I.; Okada, M.; Iwasaki, K.; Miki, H.; Takahashi, S.; Kouzmenko, A.; Nohara, K.; Chiba, T.; et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nat. Cell Biol. 2007, 446, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Ehrlund, A.; Anthonisen, E.H.; Gustafsson, N.; Venteclef, N.; Remen, K.R.; Damdimopoulos, A.E.; Galeeva, A.; Pelto-Huikko, M.; Lalli, E.; Steffensen, K.; et al. E3 Ubiquitin Ligase RNF31 Cooperates with DAX-1 in Transcriptional Repression of Steroidogenesis. Mol. Cell Biol. 2009, 29, 2230–2242. [Google Scholar] [CrossRef] [Green Version]
- Rajapurohitam, V.; Bedard, N.; Wing, S.S. Control of ubiquitination of proteins in rat tissues by ubiquitin conjugating enzymes and isopeptidases. Am. J. Physiol. Metab. 2002, 282, E739–E745. [Google Scholar] [CrossRef] [Green Version]
- An, J.Y.; Kim, E.-A.; Jiang, Y.; Zakrzewska, A.; Kim, D.E.; Lee, M.J.; Mook-Jung, I.; Zhang, Y.; Kwon, Y.T. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc. Natl. Acad. Sci. USA 2010, 107, 1912–1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickins, R.A.; Frew, I.; House, C.M.; O’Bryan, M.; Holloway, A.J.; Haviv, I.; Traficante, N.; de Kretser, D.M.; Bowtell, D.D.L. The Ubiquitin Ligase Component Siah1a Is Required for Completion of Meiosis I in Male Mice. Mol. Cell Biol. 2002, 22, 2294–2303. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Lin, C.; Kim, S.T.; Roig, I.; Chen, H.; Liu, L.; Veith, G.M.; Jin, R.U.; Keeney, S.; Jasin, M.; et al. The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 2011, 356, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Nian, H.; Zhang, W.; Shi, H.; Zhao, Q.; Xie, Q.; Liao, S.; Zhang, Y.; Zhang, Z.; Wang, C.; Han, C. Mouse RING finger protein Rnf133 is a testis-specific endoplasmic reticulum-associated E3 ubiquitin ligase. Cell Res. 2008, 18, 800–802. [Google Scholar] [CrossRef]
- Lerer-Goldshtein, T.; Bel, S.; Shpungin, S.; Pery, E.; Motro, B.; Goldstein, R.S.; Bar-Sheshet, S.I.; Breitbart, H.; Nir, U. TMF/ARA160: A key regulator of sperm development. Dev. Biol. 2010, 348, 12–21. [Google Scholar] [CrossRef] [Green Version]
- I Rodríguez, C.; Stewart, C.L. Disruption of the ubiquitin ligase HERC4 causes defects in spermatozoon maturation and impaired fertility. Dev. Biol. 2007, 312, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Shang, X.; Tian, Y.; Zhao, W.; He, Y.; Chen, K.; Cheng, H.; Zhou, R. Ubiquitin C-terminal hydrolase-L1 (Uch-L1) correlates with gonadal transformation in the rice field eel. FEBS J. 2008, 275, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Nickel, B.E.; Roth, S.Y.; Cook, R.G.; Allis, C.D.; Davie, J.R. Changes in the histone H2A variant H2A.Z and polyubiquitinated histone species in developing trout testis. Biochemistry 1987, 26, 4417–4421. [Google Scholar] [CrossRef] [PubMed]
- Redon, E.; Bosseboeuf, A.; Rocancourt, C.; Da Silva, C.; Wincker, P.; Mazan, S.; Sourdaine, P. Stage-specific gene expression during spermatogenesis in the dogfish (Scyliorhinus canicula). Reproduction 2010, 140, 57–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forne, I.; Castellana, B.; Marín-Juez, R.; Cerda, J.; Abián, J.; Planas, J.V. Transcriptional and proteomic profiling of flatfish (Solea senegalensis) spermatogenesis. Proteomics 2011, 11, 2195–2211. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, S.; Gao, F.; Meng, L.; Hu, Q.; Song, W.; Shao, C.; Lv, W. SCAR transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semiliaevis). J. Agric. Biotechnol. 2017, 22, 787–792. [Google Scholar]
- Fryburg, D.A. Insulin-like growth factor I exerts growth hormone- and insulin-like actions on human muscle protein metabolism. Am. J. Physiol. Metab. 1994, 267, E331–E336. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, X.; Sha, Z.; Yang, C.; Liu, S.; Wang, N.; Chen, S.-L. Establishment and Characterization of a Testicular Cell Line from the Half-Smooth Tongue Sole, Cynoglossus semilaevis. Int. J. Biol. Sci. 2011, 7, 452–459. [Google Scholar] [CrossRef] [Green Version]
- Sun, A.; Wang, T.Z.; Wang, N.; Liu, X.F.; Sha, Z.X.; Chen, S.L. Establishment and characterization of an ovarian cell line from half-smooth tongue sole Cynoglossus semilaevis. J. Fish Biol. 2015, 86, 46–59. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Y.; Wang, W.; Wang, Q.; Zhang, N.; Lin, F.; Wang, N.; Shao, C.; Dong, Z.; Li, Y.; et al. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 2017, 7, 42213. [Google Scholar] [CrossRef]
- Zhu, Y.; Meng, L.; Xu, W.; Cui, Z.; Zhang, N.; Guo, H.; Wang, N.; Shao, C.; Chen, S. The autosomal Gsdf gene plays a role in male gonad development in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 2018, 8, 17716. [Google Scholar] [CrossRef]
- Dong, X.; Chen, S.; Ji, X.; Shao, C. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Oceanol. Sin. 2011, 30, 68–77. [Google Scholar] [CrossRef]
- Deng, S.-P.; Chen, S.-L.; Xu, J.-Y.; Liu, B.-W. Molecular cloning, characterization and expression analysis of gonadal P450 aromatase in the half-smooth tongue-sole, Cynoglossus semilaevis. Aquaculture 2009, 287, 211–218. [Google Scholar] [CrossRef]
- Xu, W.; Li, H.; Dong, Z.; Cui, Z.; Zhang, N.; Meng, L.; Zhu, Y.; Liu, Y.; Li, Y.; Guo, H.; et al. Ubiquitin ligase gene neurl3 plays a role in spermatogenesis of half-smooth tongue sole (Cynoglossus semilaevis) by regulating testis protein ubiquitination. Gene 2016, 592, 215–220. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.-N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, W.; Zhu, Y.; Zhang, N.; Ma, J.; Sun, A.; Cui, Z.; Gao, F.; Wang, N.; Shao, C.; et al. Characterization and expression pattern of r-spondin1 in Cynoglossus semilaevis. J. Exp. Zool. Part B Mol. Dev. Evol. 2017, 328, 772–780. [Google Scholar] [CrossRef]
- Chen, S.-L.; Tian, Y.-S.; Yang, J.-F.; Shao, C.; Ji, X.-S.; Zhai, J.-M.; Liao, X.-L.; Zhuang, Z.-M.; Su, P.-Z.; Xu, J.-Y.; et al. Artificial Gynogenesis and Sex Determination in Half-Smooth Tongue Sole (Cynoglossus semilaevis). Mar. Biotechnol. 2008, 11, 243–251. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, W.; Du, X.; Zhao, J.; Liu, X.; Li, X.; Zhang, Q.; Wang, X. Sexually dimorphic expression in developing and adult gonads shows an important role of gonadal soma-derived factor during sex differentiation in olive flounder ( Paralichthys olivaceus ). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 210, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Beitel, L.K.; A Elhaji, Y.; Lumbroso, R.; Wing, S.S.; Panet-Raymond, V.; Gottlieb, B.; Pinsky, L.; A Trifiro, M. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 2002, 29, 41–60. [Google Scholar] [CrossRef]
- Morgan, C. Micro RNAs and the Sex Specific Development of the Neonatal Brain: A Point of Vulnerability to the Programming Effects of Prenatal Stress. 2015. Publicly Accessible Penn Dissertations. 1098. Available online: https://repository.upenn.edu/edissertations/1098(accessed on 11 November 2017).
- Huang, X.; Qin, Q.; Gong, K.; Wu, C.; Zhou, Y.; Chen, Q.; Feng, W.; Xing, Y.; Wang, C.; Wang, Y.; et al. Comparative analyses of the Sox9a-Amh-Cyp19a1a regulatory Cascade in Autotetraploid fish and its diploid parent. BMC Genet. 2020, 21, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Lucy, M.C. Growth hormone regulation of follicular growth. Reprod. Fertil. Dev. 2012, 24, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, M. Insulin-like Growth Factors and Fish Reproduction. Biol. Reprod. 2010, 82, 656–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puche, J.E.; Castilla-Cortázar, I. Human conditions of insulin-like growth factor-I (IGF-I) deficiency. J. Transl. Med. 2012, 10, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Primer | Sequences (5′–3′) | Purpose | Product Size |
|---|---|---|---|
| rchy1 probeF | CACCGCGGCCGCAGCAATGGGAGCAGATAG | ISH | 213 bp |
| rchy1 probeR | CACCCTCGAGGCTGATGTTGTGCGTGAA | ||
| rchy1 F | TTTACACAAGACCTGCTTTG | qPCR | 108 bp |
| rchy1 R | TCATCTATCTGCTCCCATT | ||
| figla_tv1 F | ACATAGAGAAGTTCAAACGAGCC | qPCR | 210 bp |
| figla_tv1 R | CGGTAGCAGCTTTTAGTGTGTCT | ||
| sox9 F | AAGAACCACACAGATCAAGACAGA | qPCR | 150 bp |
| sox9 R | TAGTCATACTGTGCTCTGGTGATG | ||
| sox-9-A F | GACCAAGTGTGTAATGTGACCAAG | qPCR | 227 bp |
| sox-9-A R | GCTCTTGGTGTTGTTATATCCACG | ||
| cyp19a F | GGTGAGGATGTGACCCAGTGT | qPCR | 230 bp |
| cyp19a R | ACGGGCTGAAATCGCAAG | ||
| igf1 F | GTATCTCCTGTAGCCACACCCTCT | qPCR | 137 bp |
| igf1 R | GCCTCTCTCTCCACACACAAACT | ||
| β-actin F | CCTTGGTATGGAGTCCTGTGGC | qPCR | 150 bp |
| β-actin R | TCCTTCTGCATCCTGTCGGC | ||
| scaffold68-2F | CCTAAATGATGGATGTAGATTCTGTC | Sex genotype | |
| scaffold68-2R | GATCCAGAGAAAATAAACCCAGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Zhu, Y.; Cheng, P.; Zhang, M.; Wang, N.; Cui, Z.; Wei, M.; Xu, W. A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis. Animals 2021, 11, 3265. https://doi.org/10.3390/ani11113265
Sun Y, Zhu Y, Cheng P, Zhang M, Wang N, Cui Z, Wei M, Xu W. A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis. Animals. 2021; 11(11):3265. https://doi.org/10.3390/ani11113265
Chicago/Turabian StyleSun, Yuxuan, Ying Zhu, Peng Cheng, Mengqian Zhang, Na Wang, Zhongkai Cui, Min Wei, and Wenteng Xu. 2021. "A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis" Animals 11, no. 11: 3265. https://doi.org/10.3390/ani11113265






