PD-L1/PD-1 and CTLA-4 Expression in Equine Penile Squamous Cell Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection
- confirmed penile localization of the lesions;
- and availability of >0.5 cm2 of FFPE tumor tissue evaluated on section.
2.2. DNA Extraction and EcPV2 Detection
2.3. RNA Extraction and Real-Time PCR
2.4. Western Blotting
2.5. Immunohistochemistry
3. Results
3.1. Case Selection and Histology
3.2. DNA Extraction and EcPV2 Detection
3.3. PD-1 and PD-L1 Gene Expression
3.4. Western Blotting
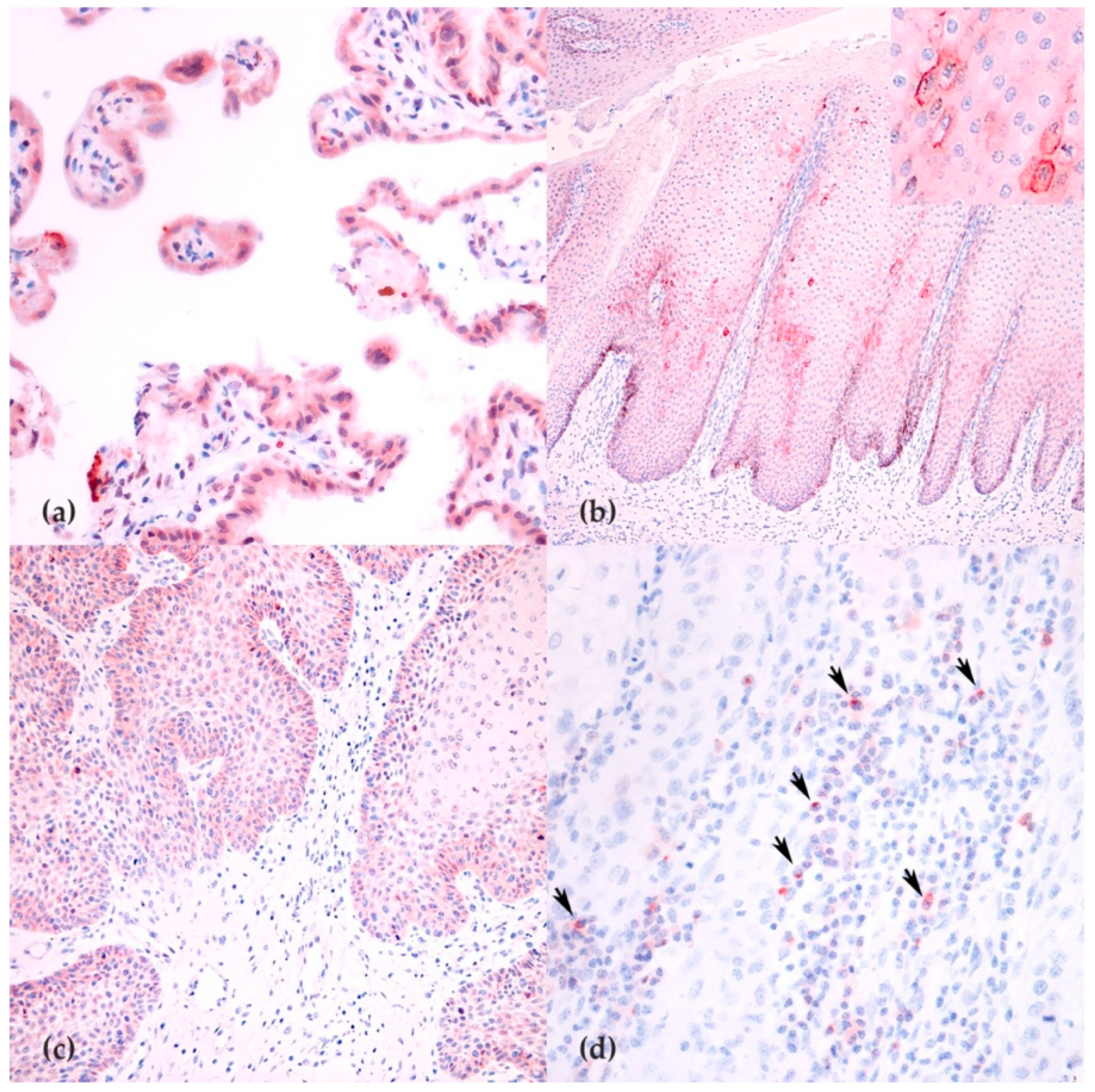
3.5. Immunohistochemistry
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van den Top, J.G.B.; Ensink, J.M.; Barneveld, A.; van Weeren, P.R. Penile and Preputial Squamous Cell Carcinoma in the Horse and Proposal of a Classification System. Equine Vet. Educ. 2011, 23, 636–648. [Google Scholar] [CrossRef]
- Ramsauer, A.S.; Wachoski-Dark, G.L.; Fraefel, C.; Tobler, K.; Brandt, S.; Knight, C.G.; Favrot, C.; Grest, P. Paving the Way for More Precise Diagnosis of EcPV2-Associated Equine Penile Lesions. BMC Vet. Res. 2019, 15, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Top, J.G.B.; Ensink, J.M.; Gröne, A.; Klein, W.R.; Barneveld, A.; van Weeren, P.R. Penile and Preputial Tumours in the Horse: Literature Review and Proposal of a Standardised Approach. Equine Vet. J. 2010, 42, 746–757. [Google Scholar] [CrossRef]
- Sykora, S.; Samek, L.; Schönthaler, K.; Palm, F.; Borzacchiello, G.; Aurich, C.; Brandt, S. EcPV-2 Is Transcriptionally Active in Equine SCC but Only Rarely Detectable in Swabs and Semen from Healthy Horses. Vet. Microbiol. 2012, 158, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Canete-Portillo, S.; Sanchez, D.F.; Cubilla, A.L. Pathology of Invasive and Intraepithelial Penile Neoplasia. Eur. Urol. Focus 2019, 5, 713–717. [Google Scholar] [CrossRef] [Green Version]
- Suárez-Bonnet, A.; Willis, C.; Pittaway, R.; Smith, K.; Mair, T.; Priestnall, S.L. Molecular Carcinogenesis in Equine Penile Cancer: A Potential Animal Model for Human Penile Cancer. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 532.e9–532.e18. [Google Scholar] [CrossRef] [Green Version]
- Porcellato, I.; Mecocci, S.; Mechelli, L.; Cappelli, K.; Brachelente, C.; Pepe, M.; Orlandi, M.; Gialletti, R.; Passeri, B.; Ferrari, A.; et al. Equine Penile Squamous Cell Carcinomas as a Model for Human Disease: A Preliminary Investigation on Tumor Immune Microenvironment. Cells 2020, 9, 2364. [Google Scholar] [CrossRef]
- Mecocci, S.; Porcellato, I.; Armando, F.; Mechelli, L.; Brachelente, C.; Pepe, M.; Gialletti, R.; Passeri, B.; Modesto, P.; Ghelardi, A.; et al. Equine Genital Squamous Cell Carcinoma Associated with EcPV2 Infection: RANKL Pathway Correlated to Inflammation and Wnt Signaling Activation. Biology 2021, 10, 244. [Google Scholar] [CrossRef]
- Petrelli, F.; Ferrara, R.; Signorelli, D.; Ghidini, A.; Proto, C.; Roudi, R.; Sabet, M.N.; Facelli, S.; Garassino, M.C.; Luciani, A.; et al. Immune Checkpoint Inhibitors and Chemotherapy in First-Line NSCLC: A Meta-Analysis. Immunotherapy 2021, 13, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Sternschuss, M.; Yerushalmi, R.; Saleh, R.R.; Amir, E.; Goldvaser, H. Efficacy and Safety of Neoadjuvant Immune Checkpoint Inhibitors in Early-Stage Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. J. Cancer Res. Clin. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jin, H.; Guo, K.; Xiang, Y.; Zhang, Y.; Du, W.; Shen, M.; Ruan, S. Results from a Meta-Analysis of Combination of PD-1/PD-L1 and CTLA-4 Inhibitors in Malignant Cancer Patients: Does PD-L1 Matter? Front. Pharmacol. 2021, 12, 217. [Google Scholar] [CrossRef]
- Högner, A.; Thuss-Patience, P. Immune Checkpoint Inhibition in Oesophago-Gastric Carcinoma. Pharmaceuticals 2021, 14, 151. [Google Scholar] [CrossRef]
- Lei, Y.; Li, X.; Huang, Q.; Zheng, X.; Liu, M. Progress and Challenges of Predictive Biomarkers for Immune Checkpoint Blockade. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 Pathway: Current Researches in Cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Zhang, L.; Zhang, M.; Xu, J.; Li, S.; Chen, Y.; Wang, W.; Yang, J.; Li, S.; Gu, M. The Role of the Programmed Cell Death Protein-1/Programmed Death-Ligand 1 Pathway, Regulatory T Cells and T Helper 17 Cells in Tumor Immunity: A Narrative Review. Ann. Transl. Med. 2020, 8, 1526. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Lu, Y.; Yang, Y.; Li, B.; Lu, P. Transcriptional and Epigenetic Regulation of PD-1 Expression. Cell. Mol. Life Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Antonangeli, F.; Natalini, A.; Garassino, M.C.; Sica, A.; Santoni, A.; Di Rosa, F. Regulation of PD-L1 Expression by NF-ΚB in Cancer. Front. Immunol. 2020, 11, 584626. [Google Scholar] [CrossRef]
- Taube, J.M.; Anders, R.A.; Young, G.D.; Xu, H.; Sharma, R.; McMiller, T.L.; Chen, S.; Klein, A.P.; Pardoll, D.M.; Topalian, S.L.; et al. Colocalization of Inflammatory Response with B7-H1 Expression in Human Melanocytic Lesions Supports an Adaptive Resistance Mechanism of Immune Escape. Sci. Transl. Med. 2012, 4, 127ra37. [Google Scholar] [CrossRef] [Green Version]
- Krähenbühl, L.; Goldinger, S.M.; Mangana, J.; Kerl, K.; Chevolet, I.; Brochez, L.; Horak, C.; Levesque, M.; Dummer, R.; Cheng, P.F. A Longitudinal Analysis of IDO and PDL1 Expression during Immune- or Targeted Therapy in Advanced Melanoma. Neoplasia 2018, 20, 218–225. [Google Scholar] [CrossRef]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, C.; Urbano, A.C.; Gameiro, A.; Ferreira, J.; Correia, J.; Ferreira, F. Serum PD-1/PD-L1 Levels, Tumor Expression and PD-L1 Somatic Mutations in HER2-Positive and Triple Negative Normal-Like Feline Mammary Carcinoma Subtypes. Cancers 2020, 12, 1386. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, N.; Konnai, S.; Takagi, S.; Kagawa, Y.; Okagawa, T.; Nishimori, A.; Ikebuchi, R.; Izumi, Y.; Deguchi, T.; Nakajima, C.; et al. A Canine Chimeric Monoclonal Antibody Targeting PD-L1 and Its Clinical Efficacy in Canine Oral Malignant Melanoma or Undifferentiated Sarcoma. Sci. Rep. 2017, 7, 8951. [Google Scholar] [CrossRef] [PubMed]
- Ottenhof, S.R.; Djajadiningrat, R.S.; de Jong, J.; Thygesen, H.H.; Horenblas, S.; Jordanova, E.S. Expression of Programmed Death Ligand 1 in Penile Cancer Is of Prognostic Value and Associated with HPV Status. J. Urol. 2017, 197, 690–697. [Google Scholar] [CrossRef]
- Ottenhof, S.R.; Djajadiningrat, R.S.; Thygesen, H.H.; Jakobs, P.J.; Józwiak, K.; Heeren, A.M.; de Jong, J.; Sanders, J.; Horenblas, S.; Jordanova, E.S. The Prognostic Value of Immune Factors in the Tumor Microenvironment of Penile Squamous Cell Carcinoma. Front. Immunol. 2018, 9, 1253. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Yao, K.; Lu, J.; Zhang, Y.; Chen, K.; Lu, J.; Zhang, C.Z.; Cao, Y. Immunophenotypes Based on the Tumor Immune Microenvironment Allow for Unsupervised Penile Cancer Patient Stratification. Cancers 2020, 12, 1796. [Google Scholar] [CrossRef] [PubMed]
- Chikuma, S. CTLA-4, an Essential Immune-Checkpoint for T-Cell Activation; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–28. [Google Scholar]
- Shi, L.; Meng, T.; Zhao, Z.; Han, J.; Zhang, W.; Gao, F.; Cai, J. CRISPR Knock out CTLA-4 Enhances the Anti-Tumor Activity of Cytotoxic T Lymphocytes. Gene 2017, 636, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Tardiel-Cyril, D.R.; Davtyan, A.; Generali, D.; Roudi, R.; Li, Y. CTLA-4 in Regulatory T Cells for Cancer Immunotherapy. Cancers 2021, 13, 1440. [Google Scholar] [CrossRef] [PubMed]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A Moving Target in Immunotherapy. Blood 2017, 131, 58–67. [Google Scholar] [CrossRef]
- Schoenfeld, J.D.; Hanna, G.J.; Jo, V.Y.; Rawal, B.; Chen, Y.-H.; Catalano, P.S.; Lako, A.; Ciantra, Z.; Weirather, J.L.; Criscitiello, S.; et al. Neoadjuvant Nivolumab or Nivolumab Plus Ipilimumab in Untreated Oral Cavity Squamous Cell Carcinoma: A Phase 2 Open-Label Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Vichi, G.; Porcellato, I.; Mechelli, L.; Fantauzzo, G.; Razzuoli, E.; Modesto, P.; Mecocci, S.; Brachelente, C. Co-Occurrence of Papillomas Related to Equus Caballus Papillomavirus Type 2 and Cutaneous Habronemiasis. Equine Vet. Educ. 2021. [Google Scholar] [CrossRef]
- Beccati, F.; Pepe, M.; Pascucci, L.; Ceccarelli, P.; Chiaradia, E.; Mancini, F.; Mandara, M.T. Sympathetic Innervation of the Suprasesamoidean Region of the Deep Digital Flexor Tendon in the Forelimbs of Horses. Vet. J. 2015, 205, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Porcellato, I.; Brachelente, C.; Cappelli, K.; Menchetti, L.; Silvestri, S.; Sforna, M.; Mecocci, S.; Iussich, S.; Leonardi, L.; Mechelli, L. FoxP3, CTLA-4, and IDO in Canine Melanocytic Tumors. Vet. Pathol. 2021, 58, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Ganbaatar, O.; Konnai, S.; Okagawa, T.; Nojima, Y.; Maekawa, N.; Minato, E.; Kobayashi, A.; Ando, R.; Sasaki, N.; Miyakoshi, D.; et al. PD-L1 Expression in Equine Malignant Melanoma and Functional Effects of PD-L1 Blockade. PLoS ONE 2020, 15, e0234218. [Google Scholar] [CrossRef]
- Benvegnen, J.; De Breuyn, B.; Gerber, V.; Rottenberg, S.; Koch, C. Immunohistochemical Analysis of Programmed Death-Ligand 1 Expression in Equine Sarcoids. J. Equine Vet. Sci. 2021, 97. [Google Scholar] [CrossRef]
- Veras, E.; Kurman, R.J.; Wang, T.-L.; Shih, I.-M. PD-L1 Expression in Human Placentas and Gestational Trophoblastic Diseases. Int. J. Gynecol. Pathol. 2017, 36, 146–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, C.A. Decade of Immune-Checkpoint Inhibitors in Cancer Therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef]
- Tagawa, M.; Yamamoto, Y.; Shimbo, G.; Iguchi, A.; Xuan, X.; Tomihari, M.; Miyahara, K. Gene and Protein Expression of a Soluble Form of CTLA-4 in a Healthy Dog. J. Vet. Med. Sci. 2017, 79, 871–875. [Google Scholar] [CrossRef]
- Urbano, A.C.; Nascimento, C.; Soares, M.; Correia, J.; Ferreira, F. Clinical Relevance of the Serum CTLA-4 in Cats with Mammary Carcinoma. Sci. Rep. 2020, 10, 3822. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Pairs Sequences | Amplicon Length | Accession and References |
|---|---|---|---|
| EcPV2-L1 | F-5′- TTGTCCAGGAGAGGGGTTAG-3′ | 81 | NC_012123 [7] |
| R-5′- TGCCTTCCTTTTCTTGGTGG-3′ | |||
| pEcPV2-L1 | FAM-CGTCCAGCACCTTCGACCACCA-TAMRA | ||
| EcPV2-E6 | F-5′-CGTTGGCCTTCTTTGCATCT-3′ | 81 | NC_012123 [8] |
| R-5′-AGGTTCAGGTCTGCTGTGTT-3′ | |||
| p-EcPV2-E6 | FAM- CCGTGTGGCTATGCTGATGACATTTGG-TAMRA | ||
| Ec-B2M DNA detection | F-5′-CTGATGTTCTCCAGGTGTTCC-3′ | 114 | NM_001082502 [7] |
| R-5′-TCAATCTCAGGCGGATGGAA-3′ | |||
| p-B2M | FAM-ACTCACGTCACCCAGCAGAGA-TAMRA | ||
| Ec-PDCD1 (PD1) | F-5′-GCCTGTGTCCTGACCACC-3′ | 150 | XM_023642815 |
| R-5′-CTCCGGGGTCTTCTCTCG-3′ | |||
| Ec-CD274 (PDL1) | F-5′-GTGCTGACTACAAGCGGATT-3′ | 120 | XM_001492842 |
| R-5′-GGTAACCCTCAGCCTGACAT-3′ | |||
| Ec-B2M cDNA | F-5′-TCCTGCTCGGGCTACTCTC-3′ | 83 | NM_001082502 [7] |
| R-5′-TGCTGGGTGACGTGAGTAAA-3′ |
| Case ID | Histological Diagnosis | DNA | cDNA | Normalized Expression | ||||
|---|---|---|---|---|---|---|---|---|
| B2M | L1 | E6 | L1 | E6 | PDCD1 | CD274 | ||
| 1 | SCC | + | +++ | ++ | + | ++ | Negative | ND |
| 2 | SCC | + | + | + | ND | ND | ND | 5.3 ± 7.3 |
| 3 | CIS | + | +++ | ++ | + | + | ND | Negative |
| 4 | SCC | + | ++++ | +++ | + | + | ND | ND |
| 6 | P | + | + | + | ND | ND | Negative | ND |
| 7 | SCC | + | + | + | ND | ND | Negative | ND |
| 8 | SCC | + | ++++ | +++ | + | + | Negative | ND |
| 9 | SCC | + | ++++ | +++ | + | + | Negative | Negative |
| 10 | SCC | + | ++++ | +++ | + | + | Negative | ND |
| 11 | SCC | + | + | + | ND | ND | Negative | ND |
| 12 | SCC | + | ++++ | +++ | + | + | Negative | ND |
| 13 | SCC | + | + | + | ND | ND | Negative | ND |
| 14 | SCC | + | ++ | + | + | ND | ND | ND |
| 15 | SCC | + | ++++ | +++++ | ++ | + | Negative | ND |
| 17 | SCC | + | ++++ | +++ | + | + | Negative | ND |
| 18 | SCC | + | ++ | ++ | ND | ND | Negative | 0.6 ± 0.7 |
| 20 | SCC | + | ND | ND | ND | ND | Negative | Negative |
| 21 | CIS | + | ++++ | +++ | ++ | ++ | ND | 9.3 ± 8.4 |
| 23 | SCC | + | +++ | ++++ | + | + | Negative | ND |
| 24 | SCC | + | ND | ND | ND | ND | Negative | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porcellato, I.; Mecocci, S.; Brachelente, C.; Cappelli, K.; Armando, F.; Tognoloni, A.; Chiaradia, E.; Stefanetti, V.; Mechelli, L.; Pepe, M.; et al. PD-L1/PD-1 and CTLA-4 Expression in Equine Penile Squamous Cell Carcinomas. Animals 2021, 11, 2121. https://doi.org/10.3390/ani11072121
Porcellato I, Mecocci S, Brachelente C, Cappelli K, Armando F, Tognoloni A, Chiaradia E, Stefanetti V, Mechelli L, Pepe M, et al. PD-L1/PD-1 and CTLA-4 Expression in Equine Penile Squamous Cell Carcinomas. Animals. 2021; 11(7):2121. https://doi.org/10.3390/ani11072121
Chicago/Turabian StylePorcellato, Ilaria, Samanta Mecocci, Chiara Brachelente, Katia Cappelli, Federico Armando, Alessia Tognoloni, Elisabetta Chiaradia, Valentina Stefanetti, Luca Mechelli, Marco Pepe, and et al. 2021. "PD-L1/PD-1 and CTLA-4 Expression in Equine Penile Squamous Cell Carcinomas" Animals 11, no. 7: 2121. https://doi.org/10.3390/ani11072121
APA StylePorcellato, I., Mecocci, S., Brachelente, C., Cappelli, K., Armando, F., Tognoloni, A., Chiaradia, E., Stefanetti, V., Mechelli, L., Pepe, M., Gialletti, R., Passeri, B., Ghelardi, A., & Razzuoli, E. (2021). PD-L1/PD-1 and CTLA-4 Expression in Equine Penile Squamous Cell Carcinomas. Animals, 11(7), 2121. https://doi.org/10.3390/ani11072121











