Investigation of Lethal Concurrent Outbreak of Chlamydiosis and Pigeon Circovirus in a Zoo
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gross and Histopathological Examinations
2.2. DNA Extraction from Fresh Tissues and Formalin-Fixed Paraffin-Embedded Tissue Blocks
2.3. Polymerase Chain Reaction Analyses of GAPDH and the 16S Ribosomal RNA (rRNA) and ompA Gene of Chlamydia psittaci
2.4. Polymerase Chain Reaction Detection of Pigeon Circovirus
2.5. Transmission Electron Microscopy (TEM)
2.6. Immunohistochemical Staining of C. psittaci
2.7. Sequence Analysis
3. Results
3.1. Gross Examination
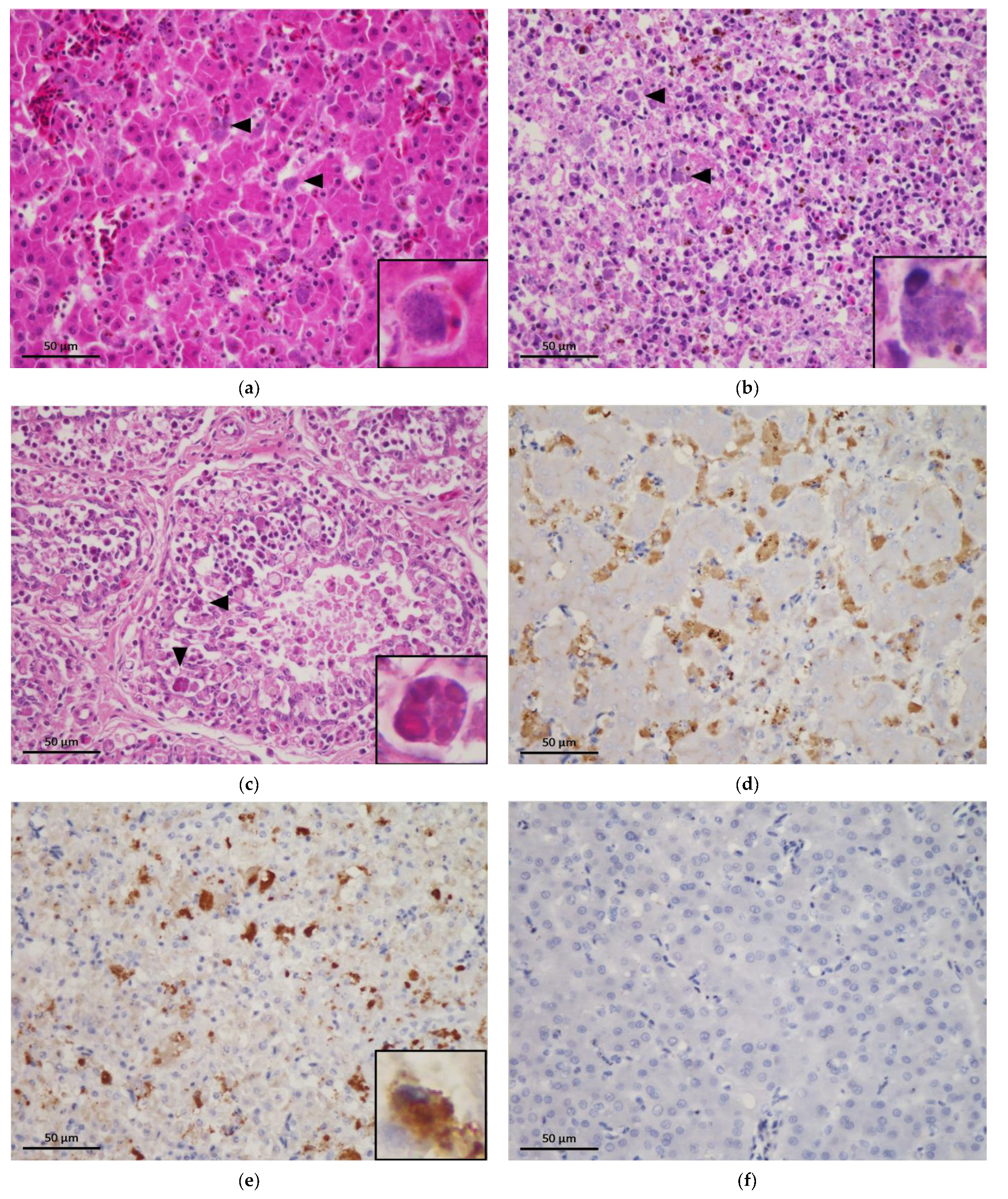
3.2. Histopathological Examination
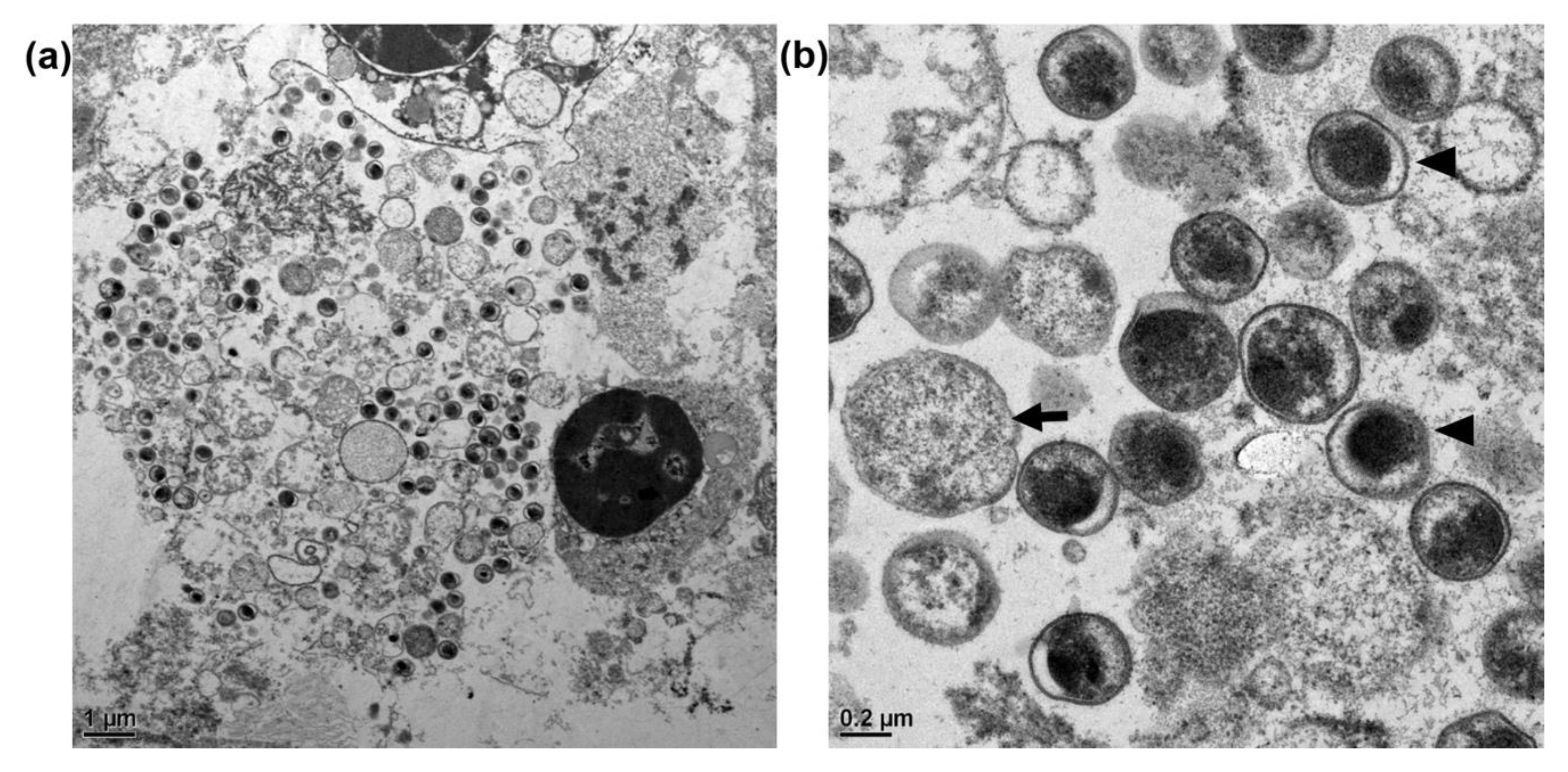
3.3. Transmission Electron Microscopy of C. psittaci
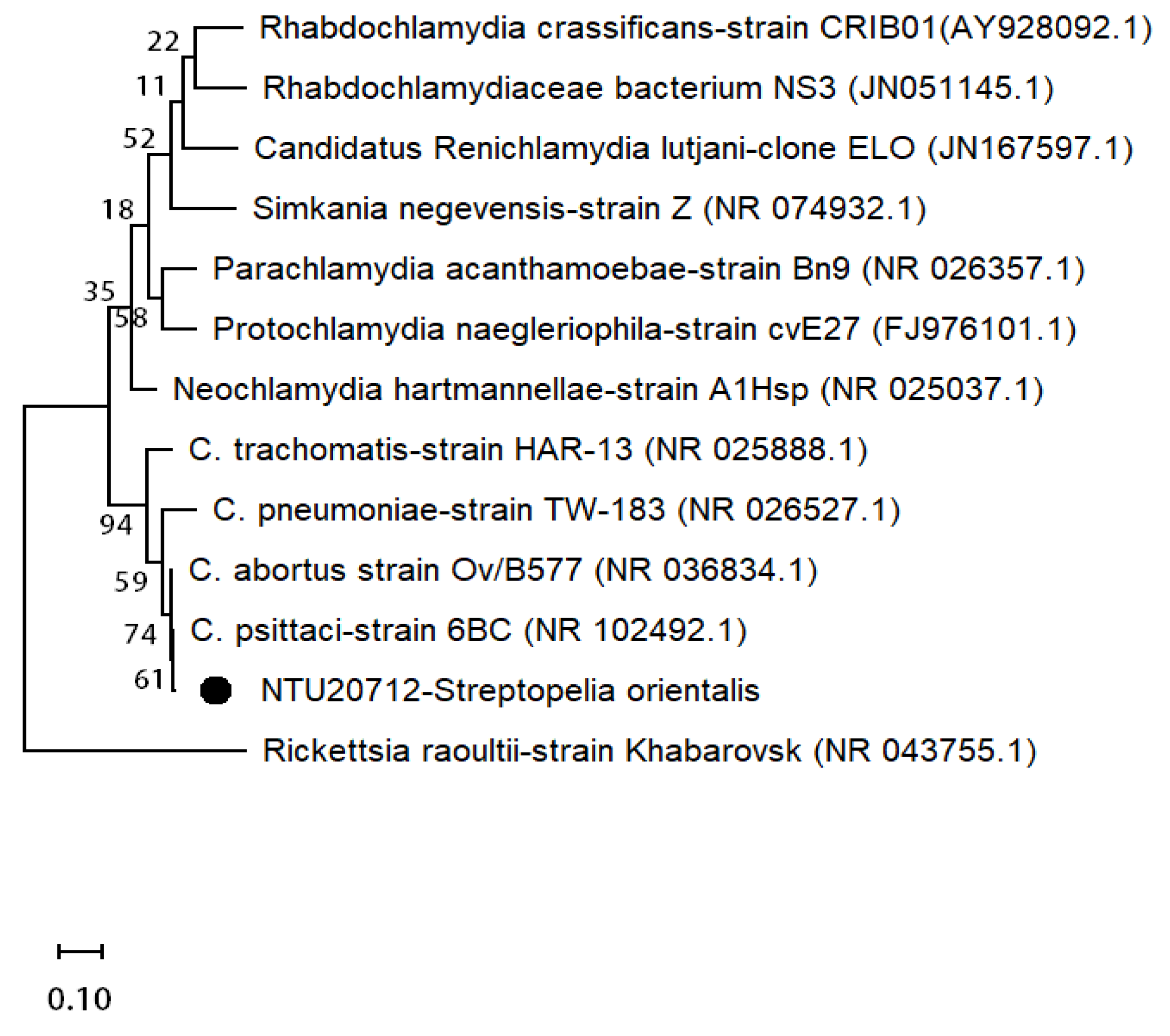
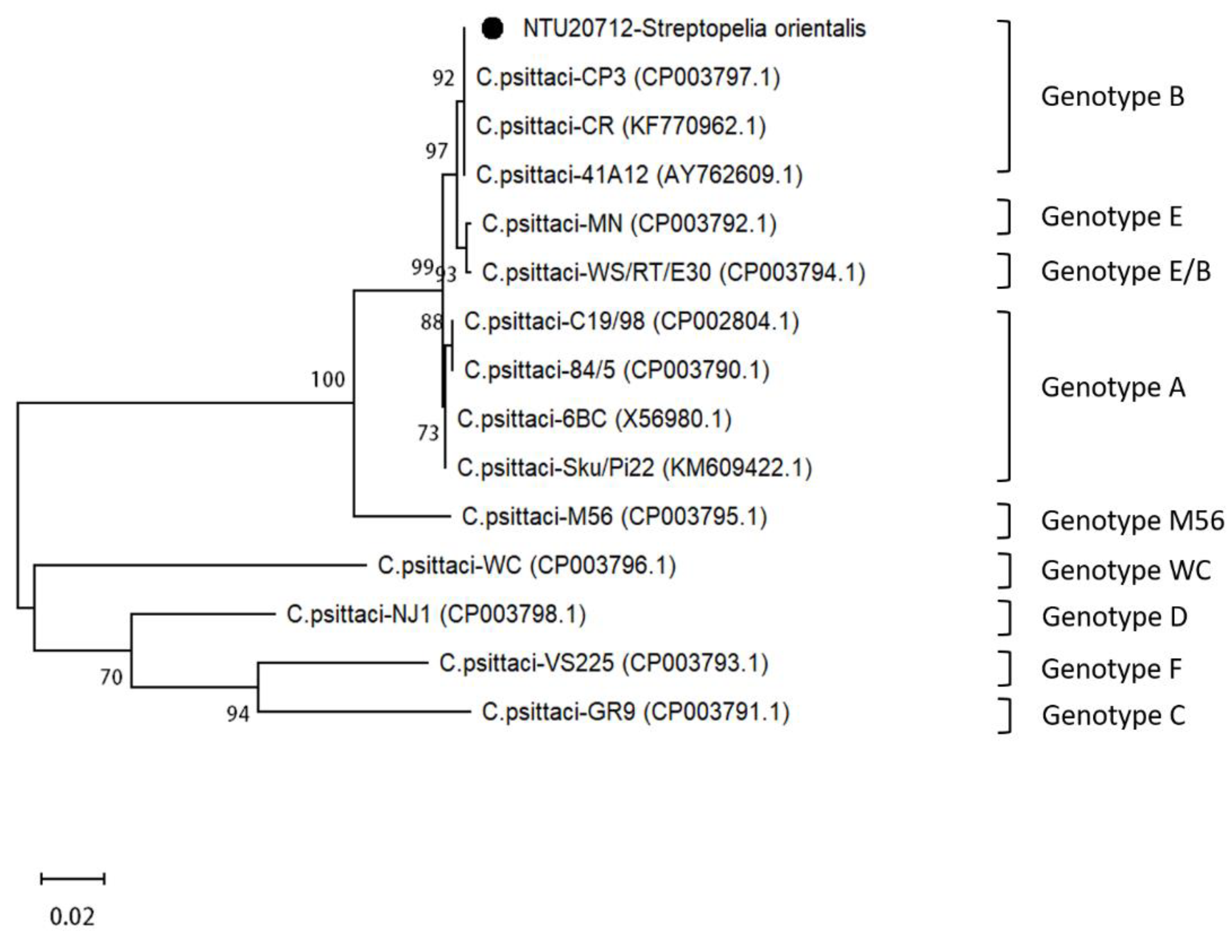
3.4. Sequence Analysis of C. psittaci
3.5. Immunohistochemical Staining of C. psittaci
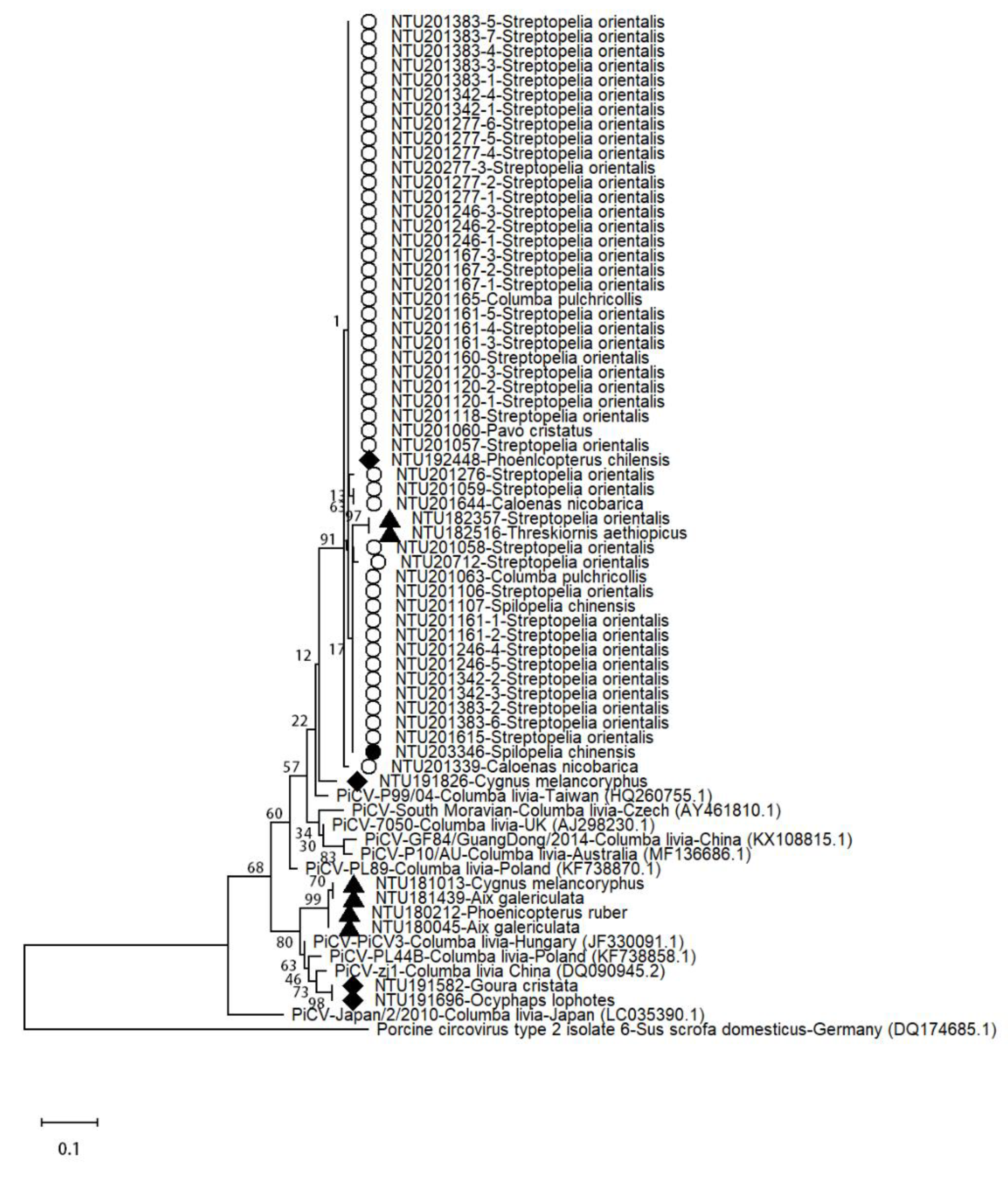
3.6. Sequence Analysis of Pigeon Circovirus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kabeya, H.; Sato, S.; Maruyama, S. Prevalence and characterization of Chlamydia DNA in zoo animals in Japan. Microbiol. Immunol. 2015, 59, 507–515. [Google Scholar] [CrossRef]
- Stenzel, T.; Pestka, D.; Choszcz, D. The prevalence and genetic characterization of Chlamydia psittaci from domestic and feral pigeons in Poland and the correlation between infection rate and incidence of pigeon circovirus. Poult. Sci. 2014, 93, 3009–3016. [Google Scholar] [CrossRef]
- Miyairi, I.; Laxton, J.D.; Wang, X.; Obert, C.A.; Arva Tatireddigari, V.R.; van Rooijen, N.; Hatch, T.P.; Byrne, G.I. Chlamydia psittaci genetic variants differ in virulence by modulation of host immunity. J. Infect. Dis. 2011, 204, 654–663. [Google Scholar] [CrossRef]
- Branley, J.; Bachmann, N.L.; Jelocnik, M.; Myers, G.S.; Polkinghorne, A. Australian human and parrot Chlamydia psittaci strains cluster within the highly virulent 6BC clade of this important zoonotic pathogen. Sci. Rep. 2016, 6, 30019. [Google Scholar] [CrossRef]
- Andersen, A.A.; Vanrompay, D. Avian chlamydiosis. Rev. Sci. Tech. 2000, 19, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Raso, T.F.; Carrasco, A.O.; Silva, J.C.; Marvulo, M.F.; Pinto, A.A. Seroprevalence of antibodies to Chlamydophila psittaci in zoo workers in Brazil. Zoonoses Public Health 2010, 57, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Dickx, V.; Van Droogenbroeck, C.; Van Vaerenbergh, B.; Herman, P.; Braeckman, L.; Vanrompay, D. Chlamydia Psittaci, Causative Agent of Avian Chlamydiosis and Human Psittacosis: Risk Assessment and Biosafety Recommendations for Laboratory use. Appl. Biosaf. 2012, 17, 82–88. [Google Scholar] [CrossRef]
- Magnino, S.; Haag-Wackernagel, D.; Geigenfeind, I.; Helmecke, S.; Dovc, A.; Prukner-Radovcic, E.; Residbegovic, E.; Ilieski, V.; Laroucau, K.; Donati, M.; et al. Chlamydial infections in feral pigeons in Europe: Review of data and focus on public health implications. Vet. Microbiol. 2009, 135, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Knittler, M.R.; Sachse, K. Chlamydia psittaci: Update on an underestimated zoonotic agent. Pathogen. Dis. 2015, 73, 1–15. [Google Scholar] [CrossRef]
- Radomski, N.; Einenkel, R.; Müller, A.; Knittler, M.R. Chlamydia–host cell interaction not only from a bird’s eye view: Some lessons from Chlamydia psittaci. FEBS Lett. 2016, 590, 3920–3940. [Google Scholar] [CrossRef]
- Vanrompay, D.; Harkinezhad, T.; van de Walle, M.; Beeckman, D.; van Droogenbroeck, C.; Verminnen, K.; Leten, R.; Martel, A.; Cauwerts, K. Chlamydophila psittaci transmission from pet birds to humans. Emerg. Infect. Dis. 2007, 13, 1108–1110. [Google Scholar] [CrossRef] [PubMed]
- Pannekoek, Y.; Dickx, V.; Beeckman, D.S.; Jolley, K.A.; Keijzers, W.C.; Vretou, E.; Maiden, M.C.; Vanrompay, D.; van der Ende, A. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS ONE 2010, 5, e14179. [Google Scholar] [CrossRef]
- Vilela, D.A.R.; Marin, S.Y.; Resende, M.; Coelho, H.L.G.; Resende, J.S.; Ferreira-Junior, F.C.; Ortiz, M.C.; Araujo, A.V.; Raso, T.F.; Martins, N.R.S. Phylogenetic analyses of Chlamydia psittaci ompA gene sequences from captive blue-fronted Amazon parrots (Amazona aestiva) with hepatic disease in Brazil. Rev. Sci. Tech. 2019, 38, 711–719. [Google Scholar] [CrossRef]
- Heidari, M.; Zhang, H.M.; Sharif, S. Marek’s Disease Virus Induces Th-2 Activity During Cytolytic Infection. Viral Immunol. 2008, 21, 203–214. [Google Scholar] [CrossRef]
- Everett, K.D.E.; Bush, R.M.; Andersen, A.A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Evol. Microbiol. 1999, 49, 415–440. [Google Scholar] [CrossRef]
- Heddema, E.R.; van Hannen, E.J.; Duim, B.; Vandenbroucke-Grauls, C.M.; Pannekoek, Y. Genotyping of Chlamydophila psittaci in human samples. Emerg. Infect. Dis. 2006, 12, 1989–1990. [Google Scholar] [CrossRef]
- Raue, R.; Schmidt, V.; Freick, M.; Reinhardt, B.; Johne, R.; Kamphausen, L.; Kaleta, E.F.; Muller, H.; Krautwald-Junghanns, M.E. A disease complex associated with pigeon circovirus infection, young pigeon disease syndrome. Avian Pathol. 2005, 34, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Liu, S.Y.; Li, K.P.; Hsieh, M.K.; Chang, P.C.; Shien, J.H.; Ou, S.C. Prevalence and Genotyping of Chlamydia psittaci from Domestic Waterfowl, Companion Birds, and Wild Birds in Taiwan. Vector-Borne Zoonotic Dis. 2019, 19, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Chen, H.; Chen, X.; Yang, X.; Yang, J.; Bavoil, P.M.; He, C. Epidemiology of Chlamydia psittaci Infection in Racing Pigeons and Pigeon Fanciers in Beijing, China. Zoonoses Public Health 2015, 62, 401–406. [Google Scholar] [CrossRef]
- Cano-Terriza, D.; Guerra, R.; Lecollinet, S.; Cerda-Cuellar, M.; Cabezon, O.; Almeria, S.; Garcia-Bocanegra, I. Epidemiological survey of zoonotic pathogens in feral pigeons (Columba livia var. domestica) and sympatric zoo species in Southern Spain. Comp. Immunol. Microbiol. Infect. Dis. 2015, 43, 22–27. [Google Scholar] [CrossRef]
- Voigt, A.; Schöfl, G.; Saluz, H.P. The Chlamydia psittaci Genome: A Comparative Analysis of Intracellular Pathogens. PLoS ONE 2012, 7, e35097. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Lagae, S.; Kalmar, I.; Borel, N.; Pospischil, A.; Vanrompay, D. Pathogenicity of low and highly virulent Chlamydia psittaci isolates for specific-pathogen-free chickens. Avian Dis. 2013, 57, 242–247. [Google Scholar] [CrossRef]
- Stenzel, T.; Koncicki, A. The epidemiology, molecular characterization and clinical pathology of circovirus infections in pigeons-current knowledge. Vet. Q. 2017, 37, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Duchatel, J.P.; Todd, D.; Smyth, J.A.; Bustin, J.C.; Vindevogel, H. Observations on detection, excretion and transmission of pigeon circovirus in adult, young and embryonic pigeons. Avian Pathol. 2006, 35, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Fringuelli, E.; Scott, A.N.; Borghmans, B.J.; Duchatel, J.P.; Shivaprasad, H.L.; Raidal, S.R.; Abadie, J.X.; Franciosini, M.P.; Smyth, J.A. Sequence comparison of pigeon circoviruses. Res. Vet. Sci. 2008, 84, 311–319. [Google Scholar] [CrossRef]
- Kubíček, O.; Taras, L. Incidence of pigeon circovirus in Euroasian collared-dove (Streptopelia decaocto) detected by nested PCR. Acta Vet. Brno 2005, 74, 361–368. [Google Scholar] [CrossRef]
| Order | Species | Sample Number | |
|---|---|---|---|
| 2 Years before the Outbreak | The Outbreak Year | ||
| Anseriformes | Aix galericulata | 2 | 0 |
| Aix sponsa | 2 | 2 | |
| Anas platyrhynchos | 4 | 1 | |
| Cygnus melancoryphus | 6 | 0 | |
| Columbiformes | Caloenas nicobarica | 1 | 3 |
| Chalcophaps indica | 0 | 1 | |
| Columba pulchricollis | 1 | 2 | |
| Ducula bicolor | 2 | 0 | |
| Goura cristata | 1 | 0 | |
| Ocyphaps lophotes | 3 | 0 | |
| Spilopelia chinensis | 0 | 2 | |
| Streptopelia orientalis | 1 | 47 | |
| Streptopelia tranquebarica | 2 | 3 | |
| Galliformes | Argusianus argus | 0 | 1 |
| Lophura nycthemera | 0 | 1 | |
| Pavo cristatus | 0 | 1 | |
| Gruiformes | Balearica pavonina | 1 | 0 |
| Balearica regulorum | 1 | 0 | |
| Gallinula chloropus | 1 | 0 | |
| Musophagiformes | Musophaga violacea | 0 | 1 |
| Pelecaniformes | Eudocimus ruber | 2 | 1 |
| Pelecanus onocrotalus | 1 | 0 | |
| Pelecanus rufescens | 4 | 0 | |
| Threskiornis aethiopicus | 2 | 0 | |
| Phoenicopteriformes | Phoenicopterus chilensis | 6 | 2 |
| Phoenicopterus ruber | 1 | 1 | |
| Time | Order | Species | C. psittaci | C. psittaci-Positive Cases | ||
|---|---|---|---|---|---|---|
| PCR | H&E | IHC | ||||
| Before outbreak | Galliformes | Pavo cristatus | 0/1 | 0/1 | NA | 0/1 |
| Phoenicopteriformes | Phoenicopterus chilensis | 0/2 | 0/2 | NA | 0/2 | |
| Phoenicopteriformes | Phoenicopterus ruber | 0/1 | 0/1 | NA | 0/1 | |
| Outbreak period | Anseriformes | Aix sponsa | 1/1 | 0/1 | 0/1 | 1/1 |
| Anseriformes | Anas platyrhynchos | 1/1 | 0/1 | 0/1 | 1/1 | |
| Columbiformes | Caloenas nicobarica | 2/2 | 1/2 | 1/2 | 2/2 | |
| Columbiformes | Chalcophaps indica | 0/1 | 0/1 | NA | 0/1 | |
| Columbiformes | Columba pulchricollis | 2/2 | 2/2 | 2/2 | 2/2 | |
| Columbiformes | Spilopelia chinensis | 2/2 | 2/2 | 2/2 | 2/2 | |
| Columbiformes | Steptopelia orientalis | 46/47 | 45/47 | 45/47 | 46/47 | |
| Columbiformes | Streptopelia tranquebarica | 3/3 | 3/3 | 3/3 | 3/3 | |
| Galliformes | Argusianus argus | 1/1 | 0/1 | 0/1 | 1/1 | |
| After outbreak | Anseriformes | Aix sponsa | 0/1 | 0/1 | NA | 0/1 |
| Columbiformes | Caloenas nicobarica | 0/1 | 0/1 | NA | 0/1 | |
| Galliformes | Lophura nycthemera | 0/1 | 0/1 | NA | 0/1 | |
| Musophagiformes | Musophaga violacea | 0/1 | 0/1 | NA | 0/1 | |
| Pelecaniformes | Eudocimus ruber | 0/1 | 0/1 | NA | 0/1 | |
| Order | Species | PiCV Positive |
|---|---|---|
| Anseriformes | Aix galericulata | 2/2 |
| Anseriformes | Cygnus melancoryphus | 2/6 |
| Columbiformes | Caloenas nicobarica | 2/4 |
| Columbiformes | Columba pulchricollis | 2/3 |
| Columbiformes | Goura cristata | 1/1 |
| Columbiformes | Ocyphaps lophotes | 1/3 |
| Columbiformes | Spilopelia chinensis | 2/2 |
| Columbiformes | Steptopelia orientalis | 43/48 |
| Galliformes | Pavo cristatus | 1/1 |
| Pelecaniformes | Threskiornis aethiopicus | 1/2 |
| Phoenicopteriformes | Phoenicopterus chilensis | 1/8 |
| Phoenicopteriformes | Phoenicopterus ruber | 1/2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.-T.; Teng, C.-A.; Shih, C.-H.; Huang, W.-H.; Jiang, Y.-F.; Chang, H.-W.; Jeng, C.-R.; Lai, Y.-H.; Guo, J.-C.; Wang, P.-J.; et al. Investigation of Lethal Concurrent Outbreak of Chlamydiosis and Pigeon Circovirus in a Zoo. Animals 2021, 11, 1654. https://doi.org/10.3390/ani11061654
Chen W-T, Teng C-A, Shih C-H, Huang W-H, Jiang Y-F, Chang H-W, Jeng C-R, Lai Y-H, Guo J-C, Wang P-J, et al. Investigation of Lethal Concurrent Outbreak of Chlamydiosis and Pigeon Circovirus in a Zoo. Animals. 2021; 11(6):1654. https://doi.org/10.3390/ani11061654
Chicago/Turabian StyleChen, Wei-Tao, Chin-Ann Teng, Cheng-Hsin Shih, Wei-Hsiang Huang, Yi-Fan Jiang, Hui-Wen Chang, Chian-Ren Jeng, Yen-Hsueh Lai, Jun-Cheng Guo, Pao-Jung Wang, and et al. 2021. "Investigation of Lethal Concurrent Outbreak of Chlamydiosis and Pigeon Circovirus in a Zoo" Animals 11, no. 6: 1654. https://doi.org/10.3390/ani11061654
APA StyleChen, W.-T., Teng, C.-A., Shih, C.-H., Huang, W.-H., Jiang, Y.-F., Chang, H.-W., Jeng, C.-R., Lai, Y.-H., Guo, J.-C., Wang, P.-J., Cheng, C.-H., & Chang, Y.-C. (2021). Investigation of Lethal Concurrent Outbreak of Chlamydiosis and Pigeon Circovirus in a Zoo. Animals, 11(6), 1654. https://doi.org/10.3390/ani11061654





