Ruminal Protozoal Populations of Angus Steers Differing in Feed Efficiency
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Design, and Rumen Content Sampling
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Sequence Analysis
2.4. Volatile Fatty Acids
2.5. Statistical Analysis
3. Results
3.1. Sequence Data, Alpha-Diversity, and Beta-Diversity of Protozoal Communities
3.2. Taxonomic Composition
3.3. Ruminal VFA Proportions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef]
- McCann, J.C.; Wiley, L.M.; Forbes, T.D.; Rouquette, F.M., Jr.; Tedeschi, L.O. Relationship between the rumen microbiome and residual feed intake-efficiency of Brahman bulls stocked on bermudagrass pastures. PLoS ONE 2014, 9, e91864. [Google Scholar] [CrossRef]
- Myer, P.R.; Smith, T.P.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C. Rumen microbiome from steers differing in feed efficiency. PLoS ONE 2015, 10, e0129174. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; Coleman, G. The Rumen Protozoa, Brock; Springer Series in Contemporary Bioscience; Springer: New York, NY, USA, 1992. [Google Scholar]
- Belanche, A.; De la Fuente, G.; Moorby, J.; Newbold, C.J. Bacterial protein degradation by different rumen protozoal groups. J. Anim. Sci. 2012, 90, 4495–4504. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; De La Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The role of ciliate protozoa in the rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef]
- Kenny, D.; Fitzsimons, C.; Waters, S.; McGee, M. Invited review: Improving feed efficiency of beef cattle—The current state of the art and future challenges. Animals 2018, 12, 1815–1826. [Google Scholar] [CrossRef]
- Myer, P.; Wells, J.; Smith, T.; Kuehn, L.; Freetly, H. Cecum microbial communities from steers differing in feed efficiency. J. Anim. Sci. 2015, 93, 5327–5340. [Google Scholar] [CrossRef] [PubMed]
- Myer, P.R.; Wells, J.E.; Smith, T.P.; Kuehn, L.A.; Freetly, H.C. Microbial community profiles of the colon from steers differing in feed efficiency. Springerplus 2015, 4, 454. [Google Scholar] [CrossRef]
- Myer, P.; Wells, J.; Smith, T.; Kuehn, L.; Freetly, H. Microbial community profiles of the jejunum from steers differing in feed efficiency. J. Anim. Sci. 2016, 94, 327–338. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Powers, J.B.; Campagna, S.R.; Seay, T.B.; Embree, M.M.; Myer, P.R. Rumen fluid metabolomics of beef steers differing in feed efficiency. Metabolomics 2020, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Krysl, L.; Hess, B. Influence of supplementation on behavior of grazing cattle. J. Anim. Sci. 1993, 71, 2546–2555. [Google Scholar] [CrossRef] [PubMed]
- Koch, R.M.; Swiger, L.A.; Chambers, D.; Gregory, K.E. Efficiency of feed use in beef cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar] [CrossRef]
- Manzano, H.P.; Kononoff, P.; Fernando, S. Evaluation of rumen bacterial composition in Holstein and Jersey cows under similar dietary conditions using different sampling methods. J. Anim. Sci. 2016, 94, 170. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004, 36, 808–813. [Google Scholar] [CrossRef]
- Ishaq, S.L.; Wright, A.-D.G. Design and validation of four new primers for next-generation sequencing to target the 18S rRNA genes of gastrointestinal ciliate protozoa. Appl. Environ. Microbiol. 2014, 80, 5515–5521. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Erwin, E.; Marco, G.; Emery, E. Volatile fatty acid analyses of blood and rumen fluid by gas chromatography. J. Dairy Sci. 1961, 44, 1768–1771. [Google Scholar] [CrossRef]
- Smith, S.B.; Gotoh, T.; Greenwood, P.L. Current situation and future prospects for global beef production: Overview of special issue. Asian-Australas. J. Anim. Sci. 2018, 31, 927–932. [Google Scholar] [CrossRef] [PubMed]
- ERS U. Cattle and Beef Sector at a Glance; United States Department of Agriculture: Washington, DC, USA, 2019.
- ERS U. Cattle & Beef Statistics & Information; United States Department of Agriculture: Washington, DC, USA, 2015.
- Montaño-Bermudez, M.; Nielsen, M.K.; Deutscher, G.H. Energy requirements for maintenance of crossbred beef cattle with different genetic potential for milk. J. Anim. Sci. 1990, 68, 2279–2288. [Google Scholar] [CrossRef]
- Coleman, G.; Sandford, D.C. The engulfment and digestion of mixed rumen bacteria and individual bacterial species by single and mixed species of rumen ciliate protozoa grown in vivo. J. Agric. Sci. 1979, 92, 729–742. [Google Scholar] [CrossRef]
- Elmqvist, T.; Folke, C.; Nyström, M.; Peterson, G.; Bengtsson, J.; Walker, B.; Norberg, J. Response diversity, ecosystem change, and resilience. Front. Ecol. Environ. 2003, 1, 488–494. [Google Scholar] [CrossRef]
- Díaz, S.; Cabido, M. Vive la différence: Plant functional diversity matters to ecosystem processes. Trends Ecol. Evol. 2001, 16, 646–655. [Google Scholar] [CrossRef]
- Jouany, J.-P. Effect of rumen protozoa on nitrogen utilization by ruminants. J. Nutr. 1996, 126 (Suppl. 4), 1335S–1346S. [Google Scholar] [CrossRef]
- Belanche, A.; de la Fuente, G.; Newbold, C.J. Effect of progressive inoculation of fauna-free sheep with holotrich protozoa and total-fauna on rumen fermentation, microbial diversity and methane emissions. FEMS Microbiol. Ecol. 2015, 91, fiu026. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Martino, C.; Powers, J.B.; Campagna, S.R.; Voy, B.H.; Donohoe, D.R.; Gaffney, J.; Embree, M.M.; Myer, P.R. Rumen bacteria and serum metabolites predictive of feed efficiency phenotypes in beef cattle. Sci. Rep. 2019, 9, 19265. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Martino, C.; Schneider, L.G.; Lefler, J.; Embree, M.M.; Myer, P.R. Temporal stability of the ruminal bacterial communities in beef steers. Sci. Rep. 2019, 9, 9522. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Jones, S.E.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J.A. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. MBio 2014, 5, e01371-14. [Google Scholar] [CrossRef] [PubMed]
- Aanderud, Z.T.; Jones, S.E.; Fierer, N.; Lennon, J.T. Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front. Microbiol. 2015, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Gobet, A.; Böer, S.I.; Huse, S.M.; Van Beusekom, J.E.E.; Quince, C.; Sogin, M.L.; Boetius, A.; Ramette, A. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 2012, 6, 542–553. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Eun, L. Effects of concentrate level on the growth of Diplodinium ciliates and cellulolytic bacteria and on the ruminal fermentation of sheep. Korean J. Anim. Sci. 1990, 32, 609–614. [Google Scholar]
- Morgavi, D.P.; Martin, C.; Jouany, J.-P.; Ranilla, M.J. Rumen protozoa and methanogenesis: Not a simple cause–effect relationship. Br. J. Nutr. 2012, 107, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Deng, Q.; Liu, Y.; Yan, T.; Li, F.; Cao, Y.; Yao, J. Dynamics of methanogenesis, ruminal fermentation and fiber digestibility in ruminants following elimination of protozoa: A meta-analysis. J. Anim. Sci. Biotechnol. 2018, 9, 89. [Google Scholar] [CrossRef]
- Ushida, K.; Kayouli, C.; De Smet, S.; Jouany, J. Effect of defaunation on protein and fibre digestion in sheep fed on ammonia-treated straw-based diets with or without maize. Br. J. Nutr. 1990, 64, 765–775. [Google Scholar] [CrossRef]
- Guan, L.L.; Nkrumah, J.D.; Basarab, J.A.; Moore, S.S. Linkage of microbial ecology to phenotype: Correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol. Lett. 2008, 288, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Nagaraja, T.; Avery, T.; Galitzer, S.; Harmon, D. Effect of ionophore antibiotics on experimentally induced lactic acidosis in cattle. Am. J. Vet. Res. 1985, 46, 2444–2452. [Google Scholar]
- Coe, M.L.; Nagaraja, T.G.; Sun, Y.D.; Wallace, N.; Towne, E.G.; Kemp, K.E.; Hutcheson, J.P. Effect of virginiamycin on ruminal fermentation in cattle during adaptation to a high concentrate diet and during an induced acidosis. J. Anim. Sci. 1999, 77, 2259–2268. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.E.; Giloteaux, L.; Orellana, R.; Williams, K.H.; Robbins, M.J.; Lovley, D.R. Methane production from protozoan endosymbionts following stimulation of microbial metabolism within subsurface sediments. Front. Microbiol. 2014, 5, 366. [Google Scholar] [CrossRef] [PubMed]
- Morgavi, D.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants. Animals 2010, 4, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
| Metric | High RFI 1 | Low RFI 1 | p-Value 2 | FDR 2,3 |
|---|---|---|---|---|
| Good’s coverage | 0.99 (0.00) | 0.99 (0.00) | 0.03 | 0.07 |
| Observed OTU (Operational taxonomic unit) | 297.93 (1.79) | 306.60 (2.71) | 0.01 | 0.03 |
| Faith’s phylogenetic diversity | 142.11 (0.44) | 144.42 (0.74) | <0.001 | 0.03 |
| Chao1 | 310.11 (2.64) | 315.01 (1.08) | 0.15 | 0.22 |
| Shannon’s diversity index | 1.86 (0.12) | 1.77 (0.21) | 0.69 | 0.69 |
| Simpson’s evenness E | 0.0074 (0.00) | 0.0079 (0.00) | 0.67 | 0.69 |
| Metric 1 | Test Statistic | p-Value |
|---|---|---|
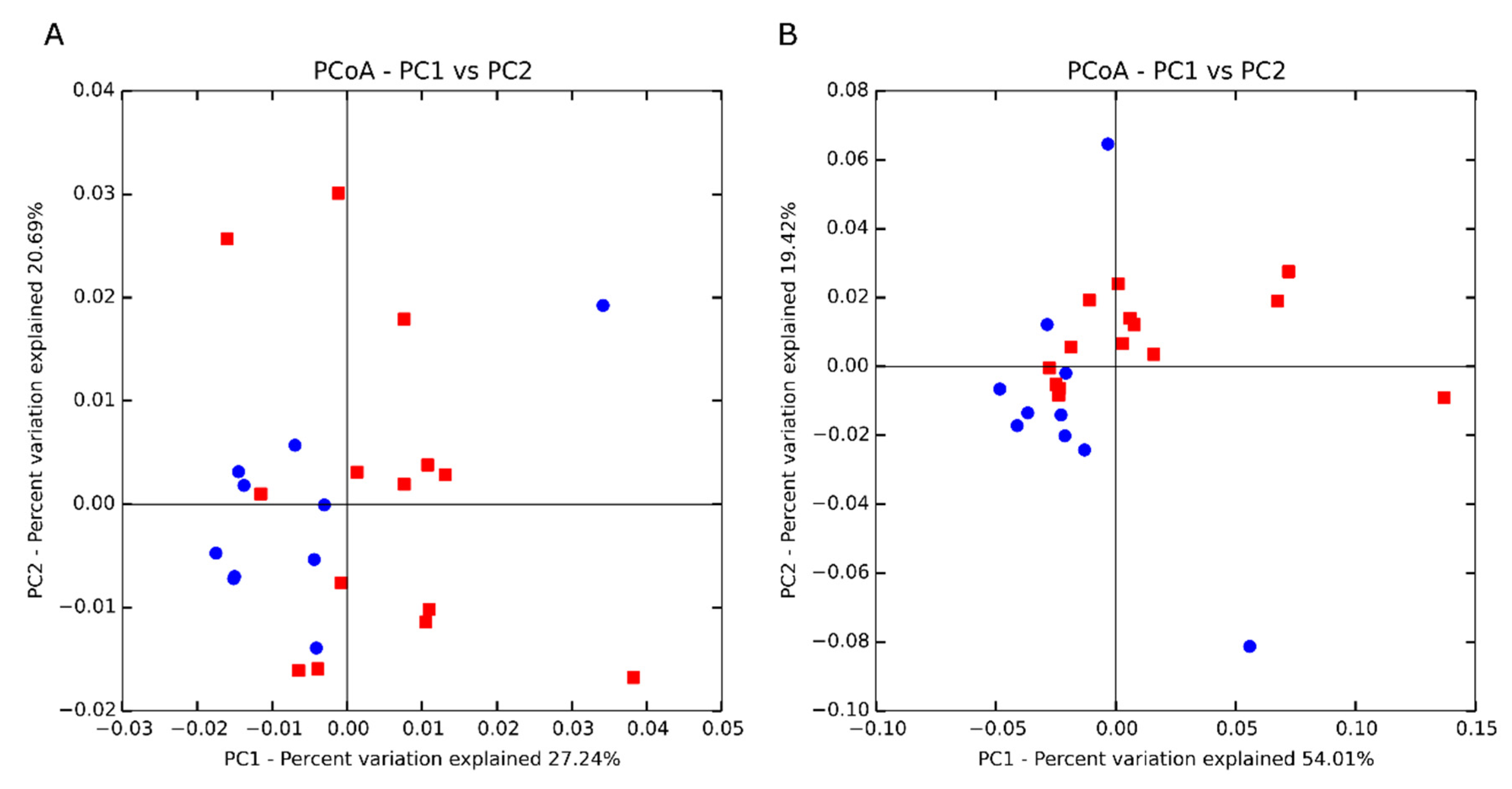
| PERMANOVA 2-weighted | 2.76 4 | 0.04 |
| PERMANOVA 2-unweighted | 0.80 4 | 0.58 |
| ANOSIM 3-weighted | 0.11 5 | 0.07 |
| ANOSIM 3-unweighted | −0.06 5 | 0.85 |
| Genus | High RFI 1 | Low RFI 1 | p-Value 2 | FDR 2,3 |
|---|---|---|---|---|
| Diplodinium | 0.12 (0.06) | 0.04 (0.03) | 0.05 4 | 0.15 4 |
| Entodinium | 0.67 (0.09) | 0.81 (0.07) | 0.13 4 | 0.19 4 |
| Isotricha | 0.06 (0.06) | 0.05 (0.05) | 0.28 4 | 0.33 4 |
| Ophryoscolex | 0.08 (0.06) | 0.02 (0.02) | 0.13 4 | 0.19 4 |
| Trichostomatia | 0.02 (0.01) | 0.05 (0.03) | 0.74 5 | 0.74 5 |
| Unassigned | 0.05 (0.00) | 0.03 (0.00) | <0.001 4 | 0.03 4 |
| VFA 1 | High RFI 2 | Low RFI 2 | p-Value 3 |
|---|---|---|---|
| Total 4 | 26.35 (2.39) | 31.49 (2.38) | 0.15 |
| Acetate | 63.88 (1.37) | 63.09 (1.33) | 0.69 |
| Propionate | 8.62 (0.89) | 7.47 (0.98) | 0.39 |
| Isobutyrate | 0.37 (0.04) | 0.42 (0.04) | 0.33 |
| Butyrate | 25.43 (1.22) | 26.01 (1.69) | 0.78 |
| Valerate | 1.69 (0.42) | 3.00 (0.88) | 0.16 |
| Taxon | Metabolite | R2 | p-Value 1,2 |
|---|---|---|---|
| Trichostomatia | Propionate | −0.41 | 0.05 |
| Ophryoscolex | Isobutyrate | −0.44 | 0.03 |
| Ophryoscolex | Butyrate | 0.43 | 0.03 |
| Diplodinium | Butyrate | 0.55 | <0.01 |
| Entodinium | Butyrate | −0.55 | <0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clemmons, B.A.; Shin, S.B.; Smith, T.P.L.; Embree, M.M.; Voy, B.H.; Schneider, L.G.; Donohoe, D.R.; McLean, K.J.; Myer, P.R. Ruminal Protozoal Populations of Angus Steers Differing in Feed Efficiency. Animals 2021, 11, 1561. https://doi.org/10.3390/ani11061561
Clemmons BA, Shin SB, Smith TPL, Embree MM, Voy BH, Schneider LG, Donohoe DR, McLean KJ, Myer PR. Ruminal Protozoal Populations of Angus Steers Differing in Feed Efficiency. Animals. 2021; 11(6):1561. https://doi.org/10.3390/ani11061561
Chicago/Turabian StyleClemmons, Brooke A., Sung B. Shin, Timothy P. L. Smith, Mallory M. Embree, Brynn H. Voy, Liesel G. Schneider, Dallas R. Donohoe, Kyle J. McLean, and Phillip R. Myer. 2021. "Ruminal Protozoal Populations of Angus Steers Differing in Feed Efficiency" Animals 11, no. 6: 1561. https://doi.org/10.3390/ani11061561
APA StyleClemmons, B. A., Shin, S. B., Smith, T. P. L., Embree, M. M., Voy, B. H., Schneider, L. G., Donohoe, D. R., McLean, K. J., & Myer, P. R. (2021). Ruminal Protozoal Populations of Angus Steers Differing in Feed Efficiency. Animals, 11(6), 1561. https://doi.org/10.3390/ani11061561






