Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
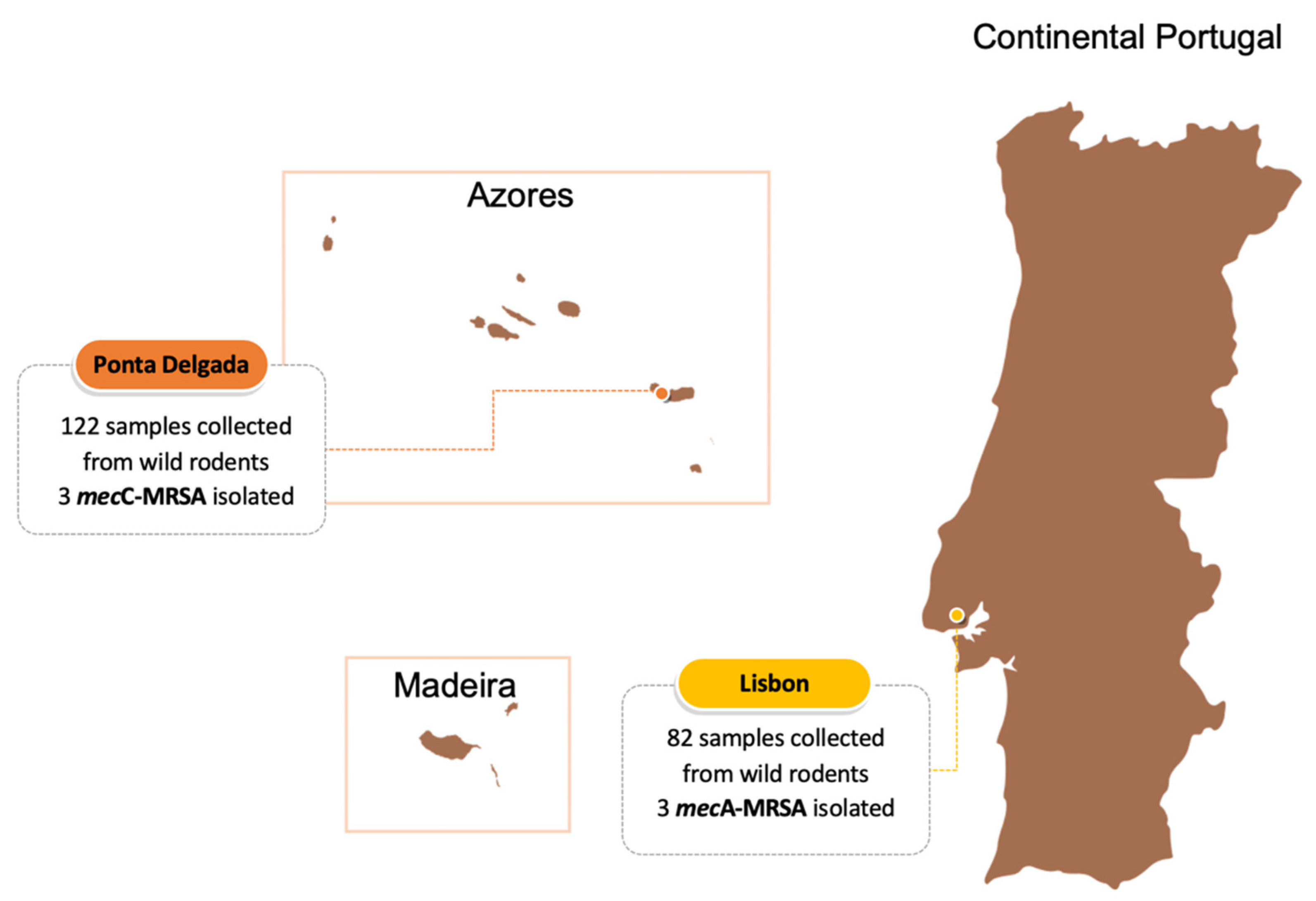
2.1. Samples and Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Antimicrobial Resistance and Virulence Genes
2.4. Molecular Typing
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Papadopoulos, P.; Papadopoulos, T.; Angelidis, A.S.; Boukouvala, E.; Zdragas, A.; Papa, A.; Hadjichristodoulou, C.; Sergelidis, D. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018, 69, 43–50. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Gavier-Widén, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; et al. Diversity of Staphylococcus aureus Isolates in European Wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef]
- Miao, J.; Chen, L.; Wang, J.; Wang, W.; Chen, D.; Li, L.; Li, B.; Deng, Y.; Xu, Z. Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 2017, 107, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Wilson, B.; Gould, I.M. Current and future treatment options for community-associated MRSA infection. Expert Opin. Pharmacother. 2018, 19, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Sieber, R.N.; Larsen, A.R.; Urth, T.R.; Iversen, S.; Møller, C.H.; Skov, R.L.; Larsen, J.; Stegger, M. Genome investigations show host adaptation and transmission of LA-MRSA CC398 from pigs into Danish healthcare institutions. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Bernier-Lachance, J.; Arsenault, J.; Usongo, V.; Parent, É.; Labrie, J.; Jacques, M.; Malouin, F.; Archambault, M. Prevalence and characteristics of Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PLoS ONE 2020, 15, e0227183. [Google Scholar] [CrossRef]
- Kadlec, K.; Entorf, M.; Peters, T. Occurrence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus in quarter milk samples from dairy cows in Germany. Front. Microbiol. 2019, 10, 1295. [Google Scholar] [CrossRef]
- Loncaric, I.; Lepuschitz, S.; Ruppitsch, W.; Trstan, A.; Andreadis, T.; Bouchlis, N.; Marbach, H.; Schauer, B.; Szostak, M.P.; Feßler, A.T.; et al. Increased genetic diversity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from companion animals. Vet. Microbiol. 2019, 235, 118–126. [Google Scholar] [CrossRef]
- Silva, V.; Capelo, J.L.; Igrejas, G.; Poeta, P. Molecular Epidemiology of Staphylococcus aureus Lineages in Wild Animals in Europe: A Review. Antibiotics 2020, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Raafat, D.; Mrochen, D.M.; Al’Sholui, F.; Heuser, E.; Ryll, R.; Pritchett-Corning, K.R.; Jacob, J.; Walther, B.; Matuschka, F.-R.; Richter, D.; et al. Molecular Epidemiology of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus in Wild, Captive and Laboratory Rats: Effect of Habitat on the Nasal S. aureus Population. Toxins 2020, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, L.; Holden, M.T.G.; Lindsay, H.; Webb, C.R.; Brown, D.F.J.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet. Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Ba, X.; Harrison, E.M.; Edwards, G.F.; Holden, M.T.G.; Larsen, A.R.; Petersen, A.; Skov, R.L.; Peacock, S.J.; Parkhill, J.; Paterson, G.K. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J. Antimicrob. Chemother. 2014, 69, 594–597. [Google Scholar] [CrossRef]
- Becker, K. Methicillin-Resistant Staphylococci and Macrococci at the Interface of Human and Animal Health. Toxins 2021, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Urushibara, N.; Aung, M.S.; Kawaguchiya, M.; Kobayashi, N. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J. Antimicrob. Chemother. 2020, 75, 46–50. [Google Scholar] [CrossRef]
- Shore, A.C.; Deasy, E.C.; Slickers, P.; Brennan, G.; O’Connell, B.; Monecke, S.; Ehricht, R.; Coleman, D.C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3765–3773. [Google Scholar] [CrossRef]
- Becker, K.; van Alen, S.; Idelevich, E.A.; Schleimer, N.; Seggewiß, J.; Mellmann, A.; Kaspar, U.; Peters, G. Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus. Emerg. Infect. Dis. 2018, 24, 242–248. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Khan, A.A.; Ali, A.; Tharmalingam, N.; Mylonakis, E.; Zahra, R. First report of mecC gene in clinical methicillin resistant S. aureus (MRSA) from tertiary care hospital Islamabad, Pakistan. J. Infect. Public Health 2020, 13, 1501–1507. [Google Scholar] [CrossRef]
- Dweba, C.C.; Zishiri, O.T.; El Zowalaty, M.E. Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus. Pathogens 2019, 8, 79. [Google Scholar] [CrossRef]
- Aklilu, E.; Chia, H.Y. First mecC and mecA Positive Livestock-Associated Methicillin Resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from Dairy Cattle in Malaysia. Microorganisms 2020, 8, 147. [Google Scholar] [CrossRef]
- Bietrix, J.; Kolenda, C.; Sapin, A.; Haenni, M.; Madec, J.-Y.; Bes, M.; Dupieux, C.; Tasse, J.; Laurent, F. Persistence and Diffusion of mecC-Positive CC130 MRSA Isolates in Dairy Farms in Meurthe-et-Moselle County (France). Front. Microbiol. 2019, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Vincze, S.; Stamm, I.; Kopp, P.A.; Hermes, J.; Adlhoch, C.; Semmler, T.; Wieler, L.H.; Lübke-Becker, A.; Walther, B. Alarming Proportions of Methicillin-Resistant Staphylococcus aureus (MRSA) in Wound Samples from Companion Animals, Germany 2010–2012. PLoS ONE 2014, 9, e85656. [Google Scholar] [CrossRef]
- Worthing, K.A.; Coombs, G.W.; Pang, S.; Abraham, S.; Saputra, S.; Trott, D.J.; Jordan, D.; Wong, H.S.; Abraham, R.J.; Norris, J.M. Isolation of mecC MRSA in Australia. J. Antimicrob. Chemother. 2016, 71, 2348–2349. [Google Scholar] [CrossRef]
- Porrero, M.C.; Harrison, E.; Fernández-Garayzábal, J.F.; Paterson, G.K.; Díez-Guerrier, A.; Holmes, M.A.; Domínguez, L. Detection of mecC-Methicillin-resistant Staphylococcus aureus isolates in river water: A potential role for water in the environmental dissemination. Environ. Microbiol. Rep. 2014, 6, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Maria de Fatima, N.F.; Penna, B.; Pereira, R.F.A.; Geraldo, R.B.; Folly, E.; Castro, H.C.; Aguiar-Alves, F. First report of meticillin-resistant Staphylococcus aureus harboring mecC gene in milk samples from cows with mastitis in southeastern Brazil. Brazilian J. Microbiol. 2020, 51, 2175–2179. [Google Scholar]
- Cuny, C.; Layer, F.; Strommenger, B.; Witte, W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS ONE 2011, 6, e24360. [Google Scholar] [CrossRef]
- Zhang, K.; Sparling, J.; Chow, B.L.; Elsayed, S.; Hussain, Z.; Church, D.L.; Gregson, D.B.; Louie, T.; Conly, J.M. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004, 42, 4947–4955. [Google Scholar] [CrossRef]
- Silva, V.; Almeida, F.; Silva, A.; Correia, S.; Carvalho, J.A.; Castro, A.P.; Ferreira, E.; Manageiro, V.; Caniça, M.; Igrejas, G.; et al. First report of linezolid-resistant cfr-positive methicillin-resistant Staphylococcus aureus in humans in Portugal. J. Glob. Antimicrob. Resist. 2019, 17, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Etienne, J.; Vandenesch, F.; Nesme, X. Relationships between Staphylococcus aureus Genetic Background, Virulence Factors, agr Groups (Alleles), and Human Disease. Infect. Immun. 2002, 70, 631–641. [Google Scholar] [CrossRef]
- van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; van Kessel, K.P.M.; van Strijp, J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Harmsen, D.; Claus, H.H.H.H.; Witte, W.; Rothgänger, J.; Claus, H.H.H.H.; Turnwald, D.; Vogel, U. Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef]
- Shopsin, B.; Mathema, B.; Alcabes, P.; Said-Salim, B.; Lina, G.; Matsuka, A.; Martinez, J.; Kreiswirth, B.N. Prevalence of agr Specificity Groups among Staphylococcus aureus Strains Colonizing Children and Their Guardians. J. Clin. Microbiol. 2003, 41, 456–459. [Google Scholar] [CrossRef]
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in Animals. Gram-Positive Pathog. 2019, 731–746. [Google Scholar] [CrossRef]
- Mrochen, D.M.; Schulz, D.; Fischer, S.; Jeske, K.; El Gohary, H.; Reil, D.; Imholt, C.; Trübe, P.; Suchomel, J.; Tricaud, E.; et al. Wild rodents and shrews are natural hosts of Staphylococcus aureus. Int. J. Med. Microbiol. 2018, 308, 590–597. [Google Scholar] [CrossRef] [PubMed]
- van de Giessen, A.W.; van Santen-Verheuvel, M.G.; Hengeveld, P.D.; Bosch, T.; Broens, E.M.; Reusken, C.B.E.M. Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev. Vet. Med. 2009, 91, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.; González-Barrio, D.; Benito, D.; García, J.T.; Viñuela, J.; Zarazaga, M.; Ruiz-Fons, F.; Torres, C. Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 2014, 69, 2061–2064. [Google Scholar] [CrossRef]
- Kmeť, V.; Čuvalová, A.; Stanko, M. Small mammals as sentinels of antimicrobial-resistant staphylococci. Folia Microbiol. (Praha) 2018, 63, 665–668. [Google Scholar] [CrossRef]
- Loncaric, I.; Kübber-Heiss, A.; Posautz, A.; Stalder, G.L.; Hoffmann, D.; Rosengarten, R.; Walzer, C. mecC-and mecA-positive meticillin-resistant Staphylococcus aureus (MRSA) isolated from livestock sharing habitat with wildlife previously tested positive for mecC-positive MRSA. Vet. Dermatol. 2014, 25, 147–148. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Gómez, P.; Alonso, C.A.; Camacho, M.C.; de la Puente, J.; Fernández-Fernández, R.; Ramiro, Y.; Quevedo, M.A.; Blanco, J.M.; Zarazaga, M.; et al. Detection of MRSA of Lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu(A) in Magpies and Cinereous Vultures in Spain. Microb. Ecol. 2019, 78, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.; Lozano, C.; González-Barrio, D.; Zarazaga, M.; Ruiz-Fons, F.; Torres, C. High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in a semi-extensive red deer (Cervus elaphus hispanicus) farm in Southern Spain. Vet. Microbiol. 2015, 177, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Kriegeskorte, A.; Ballhausen, B.; Idelevich, E.A.; Köck, R.; Friedrich, A.W.; Karch, H.; Peters, G.; Becker, K. Human MRSA isolates with novel genetic homolog, Germany. Emerg. Infect. Dis. 2012, 18, 1016–1018. [Google Scholar] [CrossRef]
- Kerschner, H.; Harrison, E.M.; Hartl, R.; Holmes, M.A.; Apfalter, P. First report of mecC MRSA in human samples from Austria: Molecular characteristics and clinical data. New Microbes New Infect. 2015, 3, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Walther, B.; Wieler, L.H.; Vincze, S.; Antão, E.-M.; Brandenburg, A.; Stamm, I.; Kopp, P.A.; Kohn, B.; Semmler, T.; Lübke-Becker, A. MRSA variant in companion animals. Emerg. Infect. Dis. 2012, 18, 2017–2020. [Google Scholar] [CrossRef] [PubMed]
- Paterson, G.K.; Harrison, E.M.; Holmes, M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014, 22, 42–47. [Google Scholar] [CrossRef]
- García-Garrote, F.; Cercenado, E.; Marín, M.; Bal, M.; Trincado, P.; Corredoira, J.; Ballesteros, C.; Pita, J.; Alonso, P.; Vindel, A. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: Emergence in Spain and report of a fatal case of bacteraemia. J. Antimicrob. Chemother. 2014, 69, 45–50. [Google Scholar] [CrossRef]
- Laurent, F.; Chardon, H.; Haenni, M.; Bes, M.; Reverdy, M.-E.; Madec, J.-Y.; Lagier, E.; Vandenesch, F.; Tristan, A. MRSA harboring mecA variant gene mecC, France. Emerg. Infect. Dis. 2012, 18, 1465. [Google Scholar] [CrossRef]
- Harrison, E.M.; Coll, F.; Toleman, M.S.; Blane, B.; Brown, N.M.; Török, M.E.; Parkhill, J.; Peacock, S.J. Genomic surveillance reveals low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus in the East of England. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Silva, V.; Almeida, F.; Carvalho, J.A.; Castro, A.P.; Ferreira, E.; Manageiro, V.; Tejedor-Junco, M.T.; Caniça, M.; Igrejas, G.; Poeta, P. Emergence of community-acquired methicillin-resistant Staphylococcus aureus EMRSA-15 clone as the predominant cause of diabetic foot ulcer infections in Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 179–186. [Google Scholar] [CrossRef]
- Silva, V.; Hermenegildo, S.; Ferreira, C.; Manaia, C.M.; Capita, R.; Alonso-Calleja, C.; Carvalho, I.; Pereira, J.E.; Maltez, L.; Capelo, J.L. Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Human Bloodstream Infections: Detection of MLSB Resistance. Antibiotics 2020, 9, 375. [Google Scholar] [CrossRef]
- Ledda, A.; Price, J.R.; Cole, K.; Llewelyn, M.J.; Kearns, A.M.; Crook, D.W.; Paul, J.; Didelot, X. Re-emergence of methicillin susceptibility in a resistant lineage of Staphylococcus aureus. J. Antimicrob. Chemother. 2017, 72, 1285–1288. [Google Scholar] [PubMed]
- Roberts, M.C.; Joshi, P.R.; Monecke, S.; Ehricht, R.; Müller, E.; Gawlik, D.; Paudel, S.; Acharya, M.; Bhattarai, S.; Pokharel, S.; et al. MRSA Strains in Nepalese Rhesus Macaques (Macaca mulatta) and Their Environment. Front. Microbiol. 2019, 10, 2505. [Google Scholar] [CrossRef]
- Gómez, P.; Lozano, C.; Camacho, M.C.; Lima-Barbero, J.-F.; Hernández, J.-M.; Zarazaga, M.; Höfle, Ú.; Torres, C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2015, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Kraushaar, B.; Fetsch, A. First description of PVL-positive methicillin-resistant Staphylococcus aureus (MRSA) in wild boar meat. Int. J. Food Microbiol. 2014, 186, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Porrero, M.C.; Mentaberre, G.; Sánchez, S.; Fernández-Llario, P.; Casas-Díaz, E.; Mateos, A.; Vidal, D.; Lavín, S.; Fernández-Garayzábal, J.-F.; Domínguez, L. Carriage of Staphylococcus aureus by Free-Living Wild Animals in Spain. Appl. Environ. Microbiol. 2014, 80, 4865–4870. [Google Scholar] [CrossRef] [PubMed]
- Seinige, D.; Von Altrock, A.; Kehrenberg, C. Genetic diversity and antibiotic susceptibility of Staphylococcus aureus isolates from wild boars. Comp. Immunol. Microbiol. Infect. Dis. 2017, 54, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; López, F.; Molina-López, R.A.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef]
- Silva, V.; Pereira, J.E.; Maltez, L.; Ferreira, E.; Manageiro, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity of methicillin-resistant staphylococci among wild Lepus granatensis: First detection of mecA-MRSA in hares. FEMS Microbiol. Ecol. 2020, 96. [Google Scholar] [CrossRef]
- Gerbig, G.R. Characterization and Whole-Genome Sequencing of Staphylococcus aureus Collected from Boston Rats. Ph.D. Thesis, Kent State University Honors College, Kent, OH, USA, 2020. [Google Scholar]
- Yang, X.; Yu, S.; Wu, Q.; Zhang, J.; Wu, S.; Rong, D. Multilocus Sequence Typing and Virulence-Associated Gene Profile Analysis of Staphylococcus aureus Isolates From Retail Ready-to-Eat Food in China. Front. Microbiol. 2018, 9, 197. [Google Scholar] [CrossRef]
- Aung, M.S.; San, T.; Urushibara, N.; San, N.; Oo, W.M.; Soe, P.E.; Kyaw, Y.; Ko, P.M.; Thu, P.P.; Hlaing, M.S.; et al. Molecular Characterization of Methicillin-Susceptible and -Resistant Staphylococcus aureus Harboring Panton-Valentine Leukocidin-Encoding Bacteriophages in a Tertiary Care Hospital in Myanmar. Microb. Drug Resist. 2019, 26, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.; Espinosa-Gongora, C.; Stamphøj, I.; Larsen, A.R.; Guardabassi, L. Carriage frequency, diversity and methicillin resistance of Staphylococcus aureus in Danish small ruminants. Vet. Microbiol. 2013, 163, 110–115. [Google Scholar] [CrossRef]
- Mama, O.M.; Ruiz-Ripa, L.; Fernández-Fernández, R.; González-Barrio, D.; Ruiz-Fons, J.F.; Torres, C. High frequency of coagulase-positive staphylococci carriage in healthy wild boar with detection of MRSA of lineage ST398-t011. FEMS Microbiol. Lett. 2019, 366, fny292. [Google Scholar] [CrossRef]
- Loncaric, I.; Kübber-Heiss, A.; Posautz, A.; Stalder, G.L.; Hoffmann, D.; Rosengarten, R.; Walzer, C. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 2013, 68, 2222–2225. [Google Scholar] [CrossRef] [PubMed]
- Gindonis, V.; Taponen, S.; Myllyniemi, A.-L.; Pyörälä, S.; Nykäsenoja, S.; Salmenlinna, S.; Lindholm, L.; Rantala, M. Occurrence and characterization of methicillin-resistant staphylococci from bovine mastitis milk samples in Finland. Acta Vet. Scand. 2013, 55, 61. [Google Scholar] [CrossRef]
- Mama, O.M.; Aspiroz, C.; Ruiz-Ripa, L.; Ceballos, S.; Iñiguez-Barrio, M.; Cercenado, E.; Azcona, J.M.; López-Cerero, L.; Seral, C.; López-Calleja, A.I.; et al. Prevalence and Genetic Characteristics of Staphylococcus aureus CC398 Isolates From Invasive Infections in Spanish Hospitals, Focusing on the Livestock-Independent CC398-MSSA Clade. Front. Microbiol. 2021, 12, 623108. [Google Scholar] [CrossRef] [PubMed]
- Porrero, M.C.; Mentaberre, G.; Sánchez, S.; Fernández-Llario, P.; Gómez-Barrero, S.; Navarro-Gonzalez, N.; Serrano, E.; Casas-Díaz, E.; Marco, I.; Fernández-Garayzabal, J.F.; et al. Methicillin resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet. J. 2013, 198, 127–130. [Google Scholar] [CrossRef]
- Akova, M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 2016, 7, 252–266. [Google Scholar] [CrossRef]
| Isolate | Host Species | Antimicrobial Resistance | Virulence Factors | Molecular Typing | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Genotype | IEC System | Other Genes | ST (CC) | spa | agr | SCCmec | ||
| VS2808 | Rattus rattus | PEN, FOX | mecC, blaZ-SCCmecXI | Type E | - | 1945 (130) | t1535 | III | XI |
| VS2809 | Rattus rattus | PEN, FOX | mecC, blaZ-SCCmecXI | Type E | - | 1945 (130) | t1535 | III | XI |
| VS2810 | Rattus norvegicus | PEN, FOX | mecC, blaZ-SCCmecXI | Type E | - | 1945 (130) | t1535 | III | XI |
| VS2811 | Rattus rattus | PEN, FOX, CIP, ERY | mecA, blaZ | - | hlb, hld | 22 (22) | t747 | I | N.T. |
| VS2812 | Rattus rattus | PEN, FOX, CIP, CN, KAN, ERY, CD | mecA, blaZ, aph(3′)-IIIa, ermA | Type A | hlb, hld | 36 (30) | t018 | I | N.T. |
| VS2813 | Rattus rattus | PEN, FOX, CIP | mecA, blaZ | - | hlb, hld | 22 (22) | t747 | I | N.T. |
| VS2814 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2815 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2816 | Rattus norvegicus | PEN | blaZ | - | hlb, hld | 1094 | t516 | I | - |
| VS2817 | Rattus rattus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2818 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2819 | Rattus rattus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2820 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2821 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2822 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2823 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2824 | Rattus norvegicus | Susceptible | - | scn | hld | 130 | t843 | III | - |
| VS2825 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2826 | Rattus rattus | Susceptible | - | - | hlb, hld | 130 | t3256 | III | - |
| VS2827 | Rattus norvegicus | Susceptible | - | - | - | 130 | t3256 | N.T. | - |
| VS2828 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1245 | t843 | III | - |
| VS2829 | Rattus rattus | Susceptible | - | - | hlb, hld | 1245 | t843 | III | - |
| VS2830 | Rattus norvegicus | PEN | blaZ | - | hld | 398 | t1451 | I | - |
| VS2831 | Rattus norvegicus | PEN | blaZ | - | hld | 398 | t1451 | I | - |
| VS2832 | Rattus norvegicus | PEN | blaZ | - | hld | 398 | t1451 | I | - |
| VS2833 | Rattus norvegicus | Susceptible | - | - | hld | 398 | t571 | I | - |
| VS2834 | Rattus rattus | Susceptible | - | Type C | hld | 5926 | t1451 | N.T. | - |
| VS2835 | Rattus norvegicus | Susceptible | - | Type C | hld | 5926 | t1451 | I | - |
| VS2836 | Rattus norvegicus | PEN | blaZ | Type E | hlb, hld | 1318 | t2078 | I | - |
| VS2837 | Rattus norvegicus | Susceptible | - | - | hld | 8 (8) | t4608 | I | - |
| VS2838 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 8 (8) | t19688 | I | - |
| VS2839 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 8 (8) | t4608 | I | - |
| VS2840 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 6574 | t1535 | III | - |
| VS2841 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 6574 | t1535 | N.T. | - |
| VS2842 | Rattus norvegicus | PEN | blaZ | - | hlb, hld | 34 (30) | t414 | N.T. | - |
| VS2843 | Rattus rattus | Susceptible | - | - | hld | 6 (5) | t16615 | I | - |
| VS2844 | Rattus rattus | Susceptible | - | - | hlb, hld | 5926 | t1451 | N.T. | - |
| VS2845 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1290 (1) | t131 | I | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, V.; Gabriel, S.I.; Borrego, S.B.; Tejedor-Junco, M.T.; Manageiro, V.; Ferreira, E.; Reis, L.; Caniça, M.; Capelo, J.L.; Igrejas, G.; et al. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals 2021, 11, 1537. https://doi.org/10.3390/ani11061537
Silva V, Gabriel SI, Borrego SB, Tejedor-Junco MT, Manageiro V, Ferreira E, Reis L, Caniça M, Capelo JL, Igrejas G, et al. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals. 2021; 11(6):1537. https://doi.org/10.3390/ani11061537
Chicago/Turabian StyleSilva, Vanessa, Sofia I. Gabriel, Sofia B. Borrego, Maria Teresa Tejedor-Junco, Vera Manageiro, Eugénia Ferreira, Lígia Reis, Manuela Caniça, José L. Capelo, Gilberto Igrejas, and et al. 2021. "Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal" Animals 11, no. 6: 1537. https://doi.org/10.3390/ani11061537
APA StyleSilva, V., Gabriel, S. I., Borrego, S. B., Tejedor-Junco, M. T., Manageiro, V., Ferreira, E., Reis, L., Caniça, M., Capelo, J. L., Igrejas, G., & Poeta, P. (2021). Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals, 11(6), 1537. https://doi.org/10.3390/ani11061537










