Comparative Analysis of Milk Microbiomes and Their Association with Bovine Mastitis in Two Farms in Central Russia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequence Library Preparation
2.3. Bioinformatics Analysis
3. Results
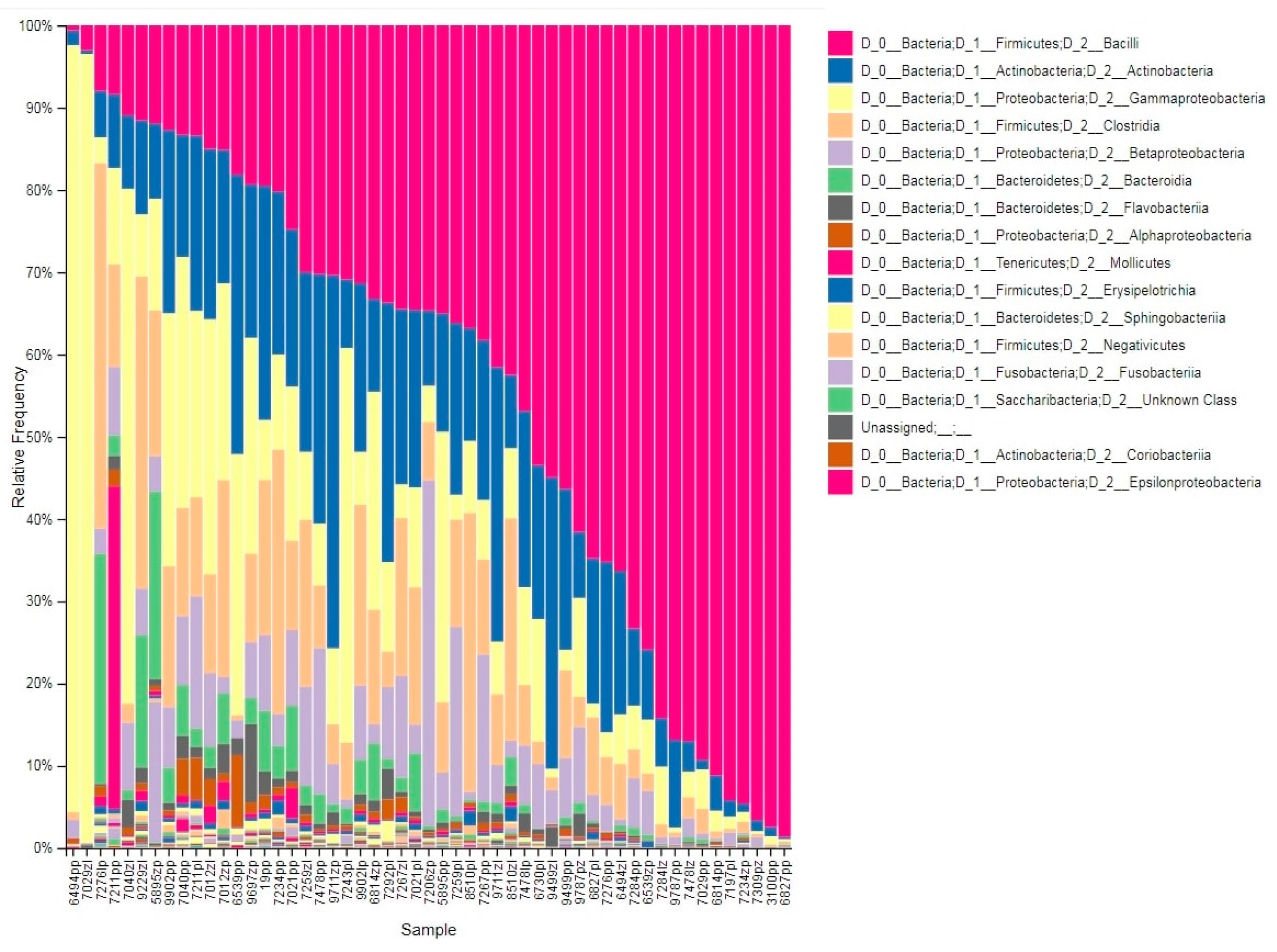
3.1. Taxonomic Profile
3.2. Diversity Analysis
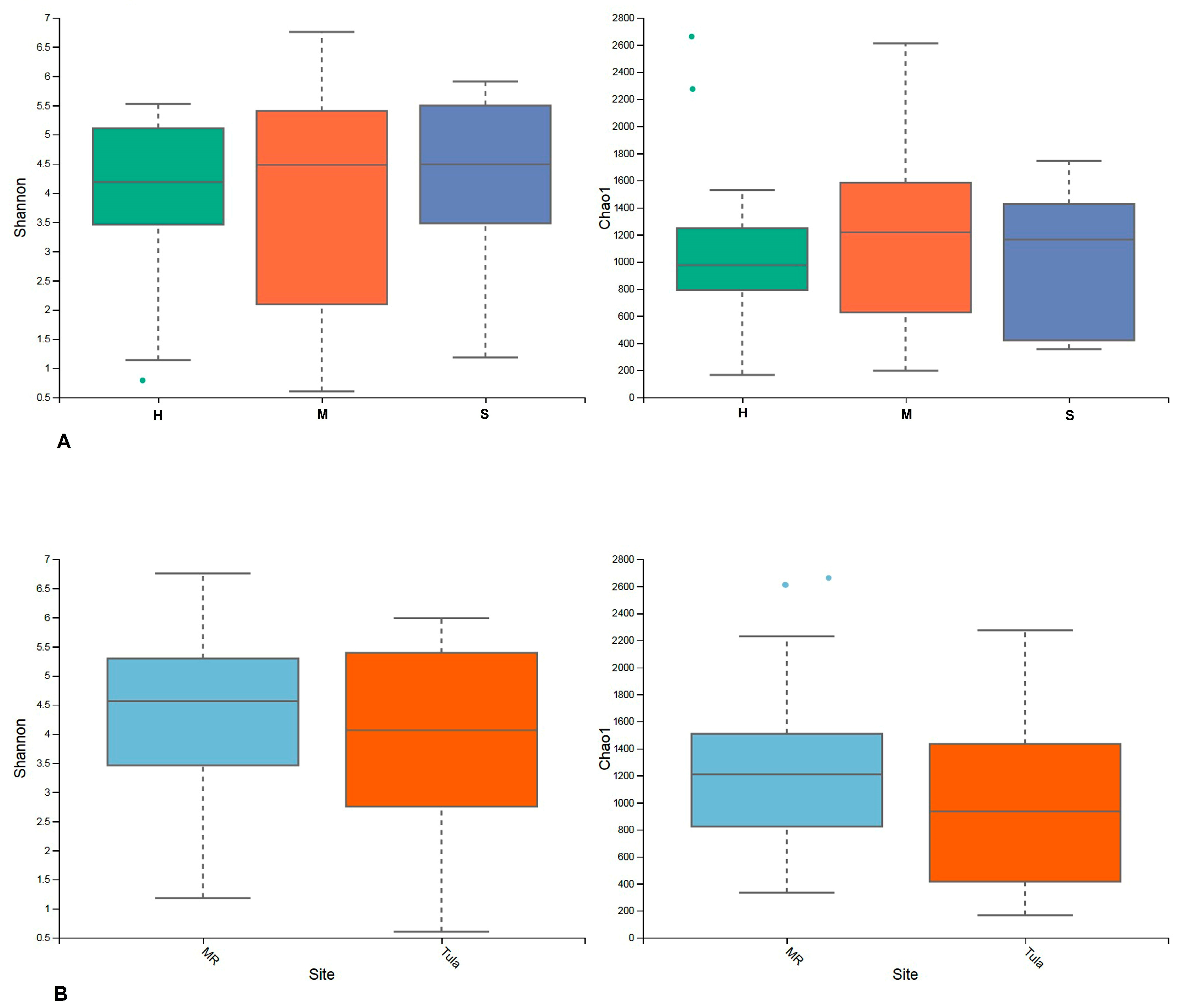
3.2.1. Alpha Diversity
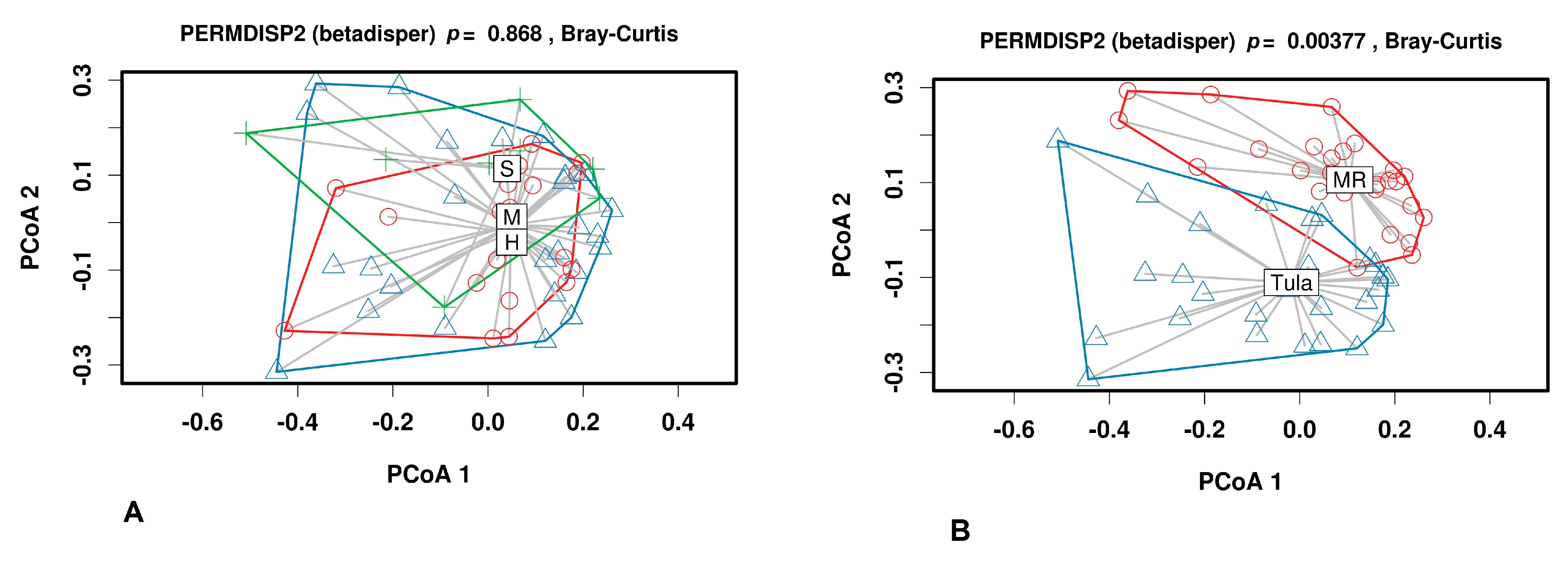
3.2.2. Beta Diversity
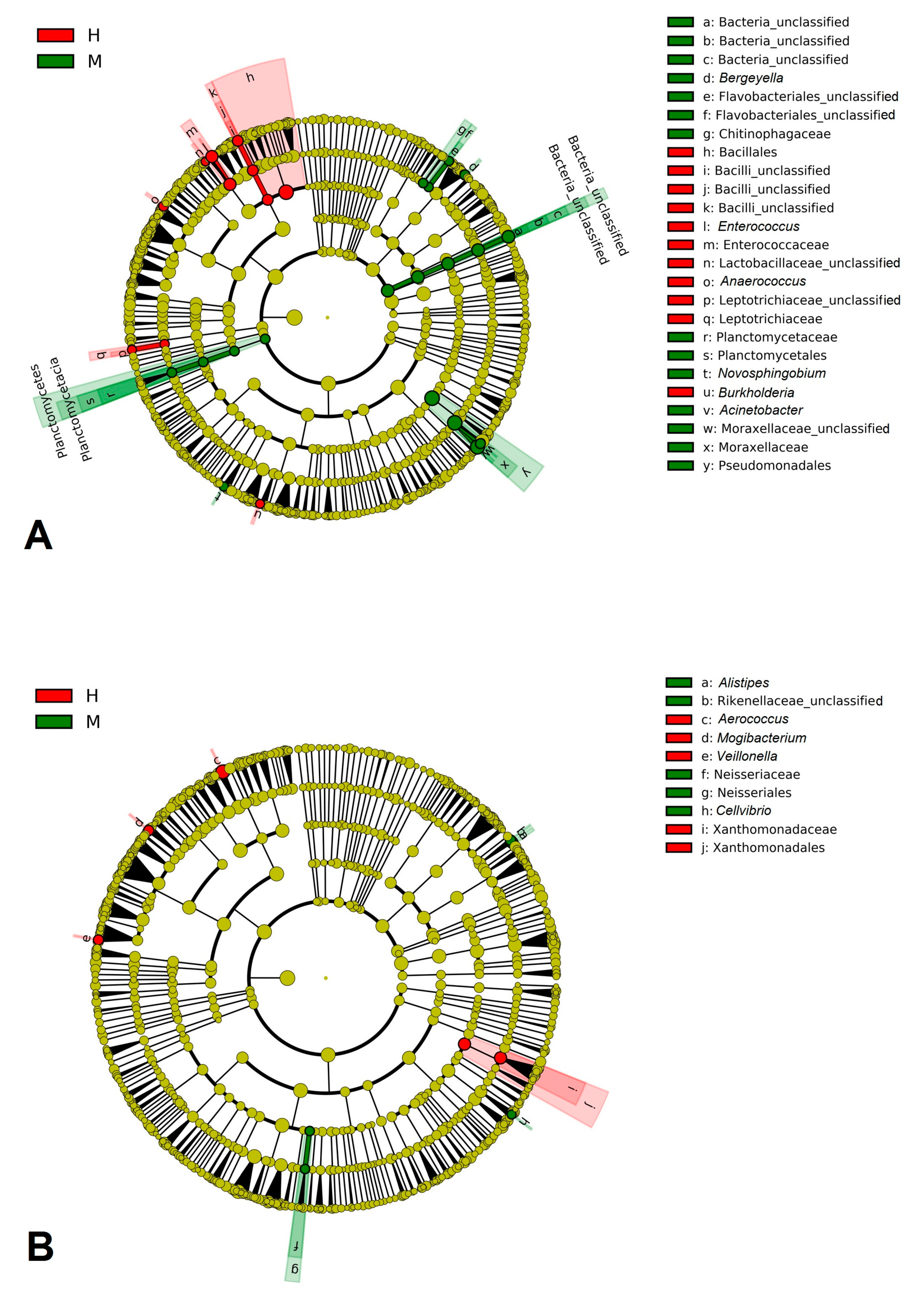
3.3. Discriminant Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heikkilä, A.M.; Nousiainen, J.; Pyorala, S. Costs of clinical mastitis with special reference to premature culling. J. Dairy Sci. 2012, 95, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Hahne, J.; Isele, D.; Berning, J.; Lipski, A. The contribution of fast growing, psychrotrophic microorganisms on biodiversity of refrigerated raw cow’s milk with high bacterial counts and their food spoilage potential. Food Microbiol. 2019, 79, 11–19. [Google Scholar] [CrossRef]
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Molecular Characterization of Staphylococcus aureus Isolated from Bovine Mastitis and Close Human Contacts in South African Dairy Herds: Genetic Diversity and Inter-Species Host Transmission. Front. Microbiol. 2017, 8, 511. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.; Pascoe, B.; Méric, G.; Mageiros, L.; Yahara, K.; Hitchings, M.D.; Friedmann, Y.; Wilkinson, T.S.; Gormley, F.J.; Mack, D.; et al. Recombination-Mediated Host Adaptation by Avian Staphylococcus aureus. Genome Biol. Evol. 2017, 9, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Islam, Z.; Espinosa-Gongora, C.; Damborg, P.; Sieber, R.N.; Munk, R.; Husted, L.; Moodley, A.; Skov, R.; Larsen, J.; Guardabassi, L. Horses in Denmark Are a Reservoir of Diverse Clones of Methicillin-Resistant and -Susceptible Staphylococcus aureus. Front. Microbiol. 2017, 8, 543. [Google Scholar] [CrossRef]
- Miragaia, M. Factors Contributing to the Evolution of MecA-Mediated β-Lactam Resistance in Staphylococci: Update and New Insights From Whole Genome Sequencing (WGS). Front Microbiol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Monaco, M.; De Araujo, F.P.; Cruciani, M.; Coccia, E.M.; Pantosti, A. Worldwide Epidemiology and Antibiotic Resistance of Staphylococcus aureus; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2016; pp. 21–56. [Google Scholar]
- Akkou, M.; Bouchiat, C.; Antri, K.; Bes, M.; Tristan, A.; Dauwalder, O.; Martins-Simoes, P.; Rasigade, J.P.; Etienne, J.; Vandenesch, F.; et al. New host shift from human to cows within Staphylococcus aureus involved in bovine mastitis and nasal carriage of animal’s caretakers. Vet. Microbiol. 2018, 223, 173–180. [Google Scholar] [CrossRef]
- Van Duijkeren, E.; Schwarz, C.; Bouchard, D.; Catry, B.; Pomba, C.; Baptiste, K.E.; A Moreno, M.; Rantala, M.; Ruzauskas, M.; Sanders, P.; et al. The use of aminoglycosides in animals within the EU: Development of resistance in animals and possible impact on human and animal health: A review. J. Antimicrob. Chemother. 2019, 74, 2480–2496. [Google Scholar] [CrossRef]
- Profet, M. The Function of Allergy: Immunological Defense Against Toxins. Q. Rev. Biol. 1991, 66, 23–62. [Google Scholar] [CrossRef]
- Asao, T.; Kumeda, Y.; Kawai, T.; Shibata, T.; Oda, H.; Haruki, K.; Nakazawa, H.; Kozaki, S. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: Estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol. Infect. 2003, 130, 33–40. [Google Scholar] [CrossRef]
- Muluk, N.B.; Altın, F.; Cingi, C. Role of Superantigens in Allergic Inflammation: Their Relationship to Allergic Rhinitis, Chronic Rhinosinusitis, Asthma, and Atopic Dermatitis. Am. J. Rhinol. Allergy 2018, 32, 502–517. [Google Scholar] [CrossRef]
- Grispoldi, L.; Massetti, L.; Sechi, P.; Iulietto, M.F.; Ceccarelli, M.; Karama, M.; Popescu, P.A.; Pandolfi, F.; Cenci-Goga, B.T. Short communication: Characterization of enterotoxin-producing Staphylococcus aureus isolated from mastitic cows. J. Dairy Sci. 2019, 102, 1059–1065. [Google Scholar] [CrossRef]
- Taponen, S.; McGuinness, D.; Hiitiö, H.; Simojoki, H.; Zadoks, R.; Pyörälä, S. Bovine milk microbiome: A more complex issue than expected. Veter Res. 2019, 50, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhang, H.; Zhang, Y.; Xiong, B.; Jiang, L. Microbiome and Metabolome Analyses of Milk From Dairy Cows With Subclinical Streptococcus agalactiae Mastitis—Potential Biomarkers. Front. Microbiol. 2019, 10, 2547. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk Microbiota: What Are We Exactly Talking About? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Herlekar, D.; Shashikant, C.; Gurjar, A.; Jayarao, B. Presence of viral and bacterial organisms in milk and their association with somatic cell counts. J. Dairy Sci. 2013, 96, 6336–6346. [Google Scholar] [CrossRef] [PubMed]
- Hertl, J.; Schukken, Y.; Welcome, F.; Tauer, L.; Gröhn, Y. Pathogen-specific effects on milk yield in repeated clinical mastitis episodes in Holstein dairy cows. J. Dairy Sci. 2014, 97, 1465–1480. [Google Scholar] [CrossRef]
- Keane, O. Symposium review: Intramammary infections—Major pathogens and strain-associated complexity. J. Dairy Sci. 2019, 102, 4713–4726. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The bovine milk microbiota: Insights and perspectives from -omics studies. Mol. BioSyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef]
- García-García, N.; Tamames, J.; Linz, A.M.; Pedrós-Alió, C.; Puente-Sánchez, F. Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. ISME J. 2019, 13, 2969–2983. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Yang, F.; Luo, J.; Wang, X.; Liu, L.; Li, H. Influences of season, parity, lactation, udder area, milk yield, and clinical symptoms on intramammary infection in dairy cows. J. Dairy Sci. 2016, 99, 6484–6493. [Google Scholar] [CrossRef] [PubMed]
- Saishu, N.; Morimoto, K.; Yamasato, H.; Ozaki, H.; Murase, T. Characterization of Aerococcus viridans isolated from milk samples from cows with mastitis and manure samples. J. Veter Med. Sci. 2015, 77, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Agriculture and Horticulture Development Board (AHDB) Knowledge Library. Available online: https://ahdb.org.uk/knowledge-library/mastitis-in-dairy-cows (accessed on 15 March 2021).
- Metzger, S.A.; Hernandez, L.L.; Suen, G.; Ruegg, P.L. Understanding the Milk Microbiota. Veter Clin. N. Am. Food Anim. Pr. 2018, 34, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Genet. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Weber, N.; Liou, D.; Dommer, J.; MacMenamin, P.; Quiñones, M.; Misner, I.; Oler, A.J.; Wan, J.; Kim, L.; McCarthy, M.C.; et al. Nephele: A cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 2017, 34, 1411–1413. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC v. 0.11.2. 2014. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 January 2020).
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Metzger, S.A.; Hernandez, L.L.; Skarlupka, J.H.; Walker, T.M.; Suen, G.; Ruegg, P.L. A Cohort Study of the Milk Microbiota of Healthy and Inflamed Bovine Mammary Glands Dryoff Through 150 Days in Milk. Front. Vet. Sci. 2018, 5, 247. [Google Scholar] [CrossRef]
- Falentin, H.; Rault, L.; Nicolas, A.; Bouchard, D.S.; Lassalas, J.; Lamberton, P.; Aubry, J.-M.; Marnet, P.G.; Le Loir, Y.; Even, S. Bovine Teat Microbiome Analysis Revealed Reduced Alpha Diversity and Significant Changes in Taxonomic Profiles in Quarters with a History of Mastitis. Front. Microbiol. 2016, 7, 480. [Google Scholar] [CrossRef]
- Oikonomou, G.; Bicalho, M.L.; Meira, E.; Rossi, R.E.; Foditsch, C.; Machado, V.S.; Teixeira, A.G.V.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbiota of Cow’s Milk; Distinguishing Healthy, Sub-Clinically and Clinically Diseased Quarters. PLoS ONE 2014, 9, e85904. [Google Scholar] [CrossRef] [PubMed]
- Dufour, S.; Dohoo, I.; Barkema, H.; Descôteaux, L.; Devries, T.; Reyher, K.; Roy, J.P.; Scholl, D. Epidemiology of coagulase-negative staphylococci intramammary infection in dairy cattle and the effect of bacteriological culture misclassification. J. Dairy Sci. 2012, 95, 3110–3124. [Google Scholar] [CrossRef]
- Leimbach, A.; Poehlein, A.; Vollmers, J.; Görlich, D.; Daniel, R.; Dobrindt, U. No evidence for a bovine mastitis Escherichia coli pathotype. BMC Genom. 2017, 18, 359. [Google Scholar] [CrossRef] [PubMed]
- Fursova, K.; Sorokin, A.; Sokolov, S.; Dzhelyadin, T.; Shulcheva, I.; Shchannikova, M.; Nikanova, D.; Artem’Eva, O.; Zinovieva, N.; Brovko, F. Virulence Factors and Phylogeny of Staphylococcus aureus Associated With Bovine Mastitis in Russia Based on Genome Sequences. Front. Vet. Sci. 2020, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Green, M.; Green, L.; Schukken, Y.; Bradley, A.; Peeler, E.; Barkema, H.; De Haas, Y.; Collis, V.; Medley, G. Somatic Cell Count Distributions During Lactation Predict Clinical Mastitis. J. Dairy Sci. 2004, 87, 1256–1264. [Google Scholar] [CrossRef]
- Grispoldi, L.; Karama, M.; Ianni, F.; La Mantia, A.; Pucciarini, L.; Camaioni, E.; Sardella, R.; Sechi, P.; Natalini, B.; Cenci-Goga, B.T. The Relationship between S. aureus and Branched-Chain Amino Acids Content in Composite Cow Milk. Animals 2019, 9, 981. [Google Scholar] [CrossRef]
- Rainard, P.; Foucras, G.; Boichard, D.; Rupp, R. Invited review: Low milk somatic cell count and susceptibility to mastitis. J. Dairy Sci. 2018, 101, 6703–6714. [Google Scholar] [CrossRef]
- Porcellato, D.; Meisal, R.; Bombelli, A.; Narvhus, J.A. A core microbiota dominates a rich microbial diversity in the bovine udder and may indicate presence of dysbiosis. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Ma, Z.; Guan, Q.; Ye, C.; Zhang, C.; Foster, J.A.; Forney, L.J. Network analysis suggests a potentially ‘evil’ alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci. Rep. 2015, 5, srep08275. [Google Scholar] [CrossRef]
- Pang, M.; Xie, X.; Bao, H.; Sun, L.; He, T.; Zhao, H.; Zhou, Y.; Zhang, L.; Zhang, H.; Wei, R.; et al. Insights Into the Bovine Milk Microbiota in Dairy Farms With Different Incidence Rates of Subclinical Mastitis. Front. Microbiol. 2018, 9, 2379. [Google Scholar] [CrossRef] [PubMed]
- Elhosseiny, N.M.; Attia, A.S. Acinetobacter: An emerging pathogen with a versatile secretome. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Klaas, I.C.; Zadoks, R.N. An update on environmental mastitis: Challenging perceptions. Transbound. Emerg. Dis. 2017, 65, 166–185. [Google Scholar] [CrossRef]
- Okello-Uma, I.; Marshall, V.M.E. Influence of mastitis on growth of starter organisms used for the manufacture of fermented milks. J. Dairy Res. 1986, 53, 631–637. [Google Scholar] [CrossRef]
- Oikonomou, G.; Machado, V.S.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbial Diversity of Bovine Mastitic Milk as Described by Pyrosequencing of Metagenomic 16s rDNA. PLoS ONE 2012, 7, e47671. [Google Scholar] [CrossRef]
- Catozzi, C.; Bonastre, A.S.; Francino, O.; Lecchi, C.; De Carlo, E.; Vecchio, D.; Martucciello, A.; Fraulo, P.; Bronzo, V.; Cuscó, A.; et al. The microbiota of water buffalo milk during mastitis. PLoS ONE 2017, 12, e0184710. [Google Scholar] [CrossRef] [PubMed]
- Polveiro, R.C.; Vidigal, P.M.P.; Mendes, T.A.D.O.; Yamatogi, R.S.; Lima, M.C.; Moreira, M.A.S. Effects of enrofloxacin treatment on the bacterial microbiota of milk from goats with persistent mastitis. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Song, L.J.; Li, L.X.; Liu, T.; Zhang, M.M.; Li, Z.; Wang, P.; Li, M.; Zuo, X.L. Fusobacterium nucleatum Causes Microbial Dysbiosis and Exacerbates Visceral Hypersensitivity in a Colonization-Independent Manner. Front. Microbiol. 2020, 11, 1281. [Google Scholar] [CrossRef] [PubMed]
- Jimbo, S.; Suleman, M.; Maina, T.; Prysliak, T.; Mulongo, M.; Perez-Casal, J. Effect of Mycoplasma bovis on bovine neutrophils. Vet. Immunol. Immunopathol. 2017, 188, 27–33. [Google Scholar] [CrossRef]
- Al-Farha, A.A.B.; Hemmatzadeh, F.; Khazandi, M.; Hoare, A.; Petrovski, K. Evaluation of effects of Mycoplasma mastitis on milk composition in dairy cattle from South Australia. BMC Vet. Res. 2017, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Qadri; Ganguly, S.; Para, P.; Praveen, P.; Wakchaure. Summer Mastitis in Cattle: A Review. J. Biol. Chem. Res. 2015, 32, 1006–1009. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokolov, S.; Fursova, K.; Shulcheva, I.; Nikanova, D.; Artyemieva, O.; Kolodina, E.; Sorokin, A.; Dzhelyadin, T.; Shchannikova, M.; Shepelyakovskaya, A.; et al. Comparative Analysis of Milk Microbiomes and Their Association with Bovine Mastitis in Two Farms in Central Russia. Animals 2021, 11, 1401. https://doi.org/10.3390/ani11051401
Sokolov S, Fursova K, Shulcheva I, Nikanova D, Artyemieva O, Kolodina E, Sorokin A, Dzhelyadin T, Shchannikova M, Shepelyakovskaya A, et al. Comparative Analysis of Milk Microbiomes and Their Association with Bovine Mastitis in Two Farms in Central Russia. Animals. 2021; 11(5):1401. https://doi.org/10.3390/ani11051401
Chicago/Turabian StyleSokolov, Sergei, Ksenia Fursova, Irina Shulcheva, Daria Nikanova, Olga Artyemieva, Evgenia Kolodina, Anatoly Sorokin, Timur Dzhelyadin, Margarita Shchannikova, Anna Shepelyakovskaya, and et al. 2021. "Comparative Analysis of Milk Microbiomes and Their Association with Bovine Mastitis in Two Farms in Central Russia" Animals 11, no. 5: 1401. https://doi.org/10.3390/ani11051401
APA StyleSokolov, S., Fursova, K., Shulcheva, I., Nikanova, D., Artyemieva, O., Kolodina, E., Sorokin, A., Dzhelyadin, T., Shchannikova, M., Shepelyakovskaya, A., Zinovieva, N., & Brovko, F. (2021). Comparative Analysis of Milk Microbiomes and Their Association with Bovine Mastitis in Two Farms in Central Russia. Animals, 11(5), 1401. https://doi.org/10.3390/ani11051401








