Genetic Heterogeneity among Chicken Infectious Anemia Viruses Detected in Italian Fowl
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
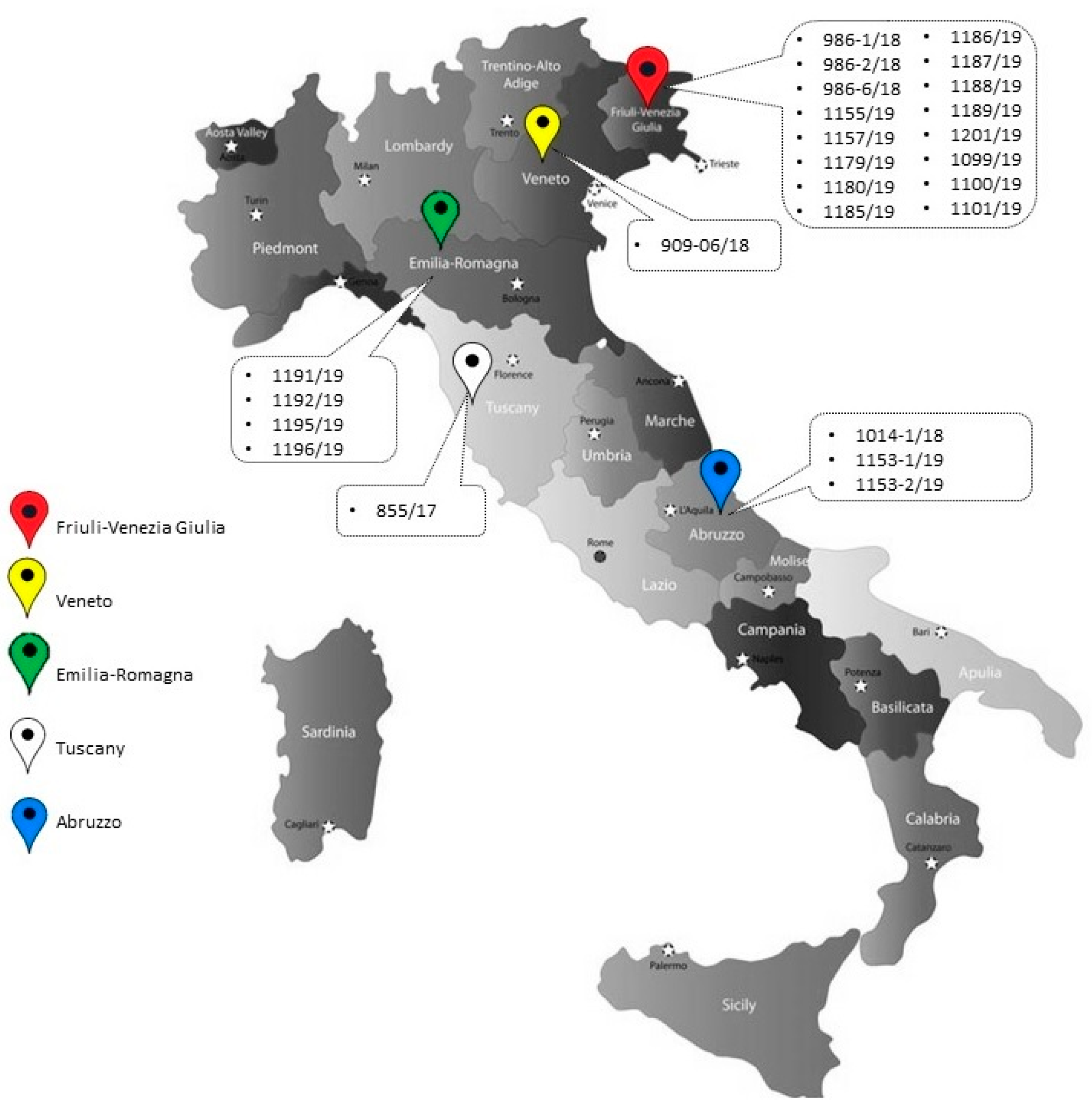
2.1. Samples
2.2. Sample Processing and DNA Extraction
2.3. Nested-PCR
2.4. Whole Genome Amplification and Sequencing
2.5. Sequence and Phylogenetic Analysis
2.6. Recombination Analysis
2.7. Accession Numbers
3. Results
3.1. Phylogenetic Analysis
3.2. Sequence Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rosario, K.; Breitbart, M.; Harrach, B.; Segalés, J.; Delwart, E.; Biagini, P.; Varsani, A. Revisiting the taxonomy of the family Circoviridae: Establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017, 162, 1447–1463. [Google Scholar] [CrossRef]
- Yuasa, N.; Taniguchi, T.; Yoshida, I. Isolation and Some Characteristics of an Agent Inducing Anemia in Chicks. Avian Dis. 1979, 23, 366. [Google Scholar] [CrossRef]
- Noteborn, M.H.M.; De Boer, G.F.; Van Roozelaar, D.J.; Karreman, C.; Kranenburg, O.; Vos, J.G.; Jeurissen, S.H.M.; Hoeben, R.C.; Zantema, A.; Koch, G.; et al. Characterization of Cloned Chicken Anemia Virus DNA That Contains All Elements for the Infectious Replication Cycle. J. Virol. 1991, 65, 3131–3139. [Google Scholar] [CrossRef] [PubMed]
- Meehan, B.M.; Todd, D.; Creelan, J.L.; Earle, J.A.P.; Hoey, E.M.; McNulty, M.S. Characterization of viral DNAs from cells infected with chicken anaemia agent: Sequence analysis of the cloned replicative form and transfection capabilities of cloned genome fragments. Arch. Virol. 1992, 124, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Lai, G.-H.; Lin, M.-K.; Lien, Y.-Y.; Fu, J.-H.; Chen, H.-J.; Huang, C.-H.; Tzen, J.T.; Lee, M.-S. Expression and characterization of highly antigenic domains of chicken anemia virus viral VP2 and VP3 subunit proteins in a recombinant E. coli for sero-diagnostic applications. BMC Vet. Res. 2013, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Creelan, J.L.; Mackie, D.P.; Rixon, F.; McNulty, M.S. Purification and biochemical characterization of chicken anaemia agent. J. Gen. Virol. 1990, 71, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Noteborn, M.H.; Van Der Eb, A.J.; Verschueren, C.A.; Koch, G. Simultaneous expression of recombinant baculovirus-encoded chicken anaemia virus (CAV) proteins VP1 and VP2 is required for formation of the CAV-specific neutralizing epitope. J. Gen. Virol. 1998, 79, 3073–3077. [Google Scholar] [CrossRef][Green Version]
- Peters, M.A.; Jackson, D.C.; Crabb, B.S.; Browning, G.F. Chicken Anemia Virus VP2 Is a Novel Dual Specificity Protein Phosphatase. J. Biol. Chem. 2002, 277, 39566–39573. [Google Scholar] [CrossRef]
- Noteborn, M.H.; Todd, D.; Verschueren, C.A.; De Gauw, H.W.; Curran, W.L.; Veldkamp, S.; Douglas, A.J.; McNulty, M.S.; Van Der Eb, A.J.; Koch, G. A single chicken anemia virus protein induces apoptosis. J. Virol. 1994, 68, 346–351. [Google Scholar] [CrossRef]
- Noteborn, M.H.M. Chicken anemia virus induced apoptosis: Underlying molecular mechanisms. Vet. Microbiol. 2004, 98, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, R.W.; Soiné, C.; Weinkle, T.; O’Connell, P.H.; Ohashi, K.; Watson, S.; Lucio, B.; Harrington, S.; Schat, K.A. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J. Virol. 1996, 70, 8872–8878. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Imada, T.; Kaji, N.; Mase, M.; Tsukamoto, K.; Tanimura, N.; Yuasa, N. Identification of a genetic determinant of pathogenicity in chicken anaemia virus. J. Gen. Virol. 2001, 82, 1233–1238. [Google Scholar] [CrossRef]
- Chowdhury, S.; Omar, A.; Aini, I.; Hair-Bejo, M.; Jamaluddin, A.; Md-Zain, B.; Kono, Y. Pathogenicity, sequence and phylogenetic analysis of Malaysian Chicken anaemia virus obtained after low and high passages in MSB-1 cells. Arch. Virol. 2003, 148, 2437–2448. [Google Scholar] [CrossRef] [PubMed]
- Natesan, S.; Kataria, J.; Dhama, K.; Rahul, S.; Baradhwaj, N. Biological and molecular characterization of chicken anaemia virus isolates of Indian origin. Virus Res. 2006, 118, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Hailemariam, Z.; Omar, A.R.; Hair-Bejo, M.; Giap, T.C. Detection and characterization of chicken anemia virus from commercial broiler breeder chickens. Virol. J. 2008, 5, 128. [Google Scholar] [CrossRef]
- Farkas, T.; Tanaka, A.; Kai, K.; Kanoe, M. Cloning and Sequencing of the Genome of Chicken Anaemia Virus (CAV) TK-5803 Strain and Comparison with Other CAV Strains. J. Vet. Med. Sci. 1996, 58, 681–684. [Google Scholar] [CrossRef][Green Version]
- Schat, K.A.; van Santen, V.L. Chicken infectious anemia virus and other Circovirus infections. In Diseases of Poultry; Saif, Y.M., Glisson, J.R., McDougald, L.R., Nolan, L.K., Swayne, D.E., Eds.; Wiley-Blackwell: Ames, IA, USA, 2008; pp. 211–235. [Google Scholar]
- Ducatez, M.F.; Owoade, A.A.; Abiola, J.O.; Muller, C.P. Molecular epidemiology of chicken anemia virus in Nigeria. Arch. Virol. 2005, 151, 97–111. [Google Scholar] [CrossRef]
- Ou, S.-C.; Lin, H.-L.; Liu, P.-C.; Huang, H.-J.; Lee, M.-S.; Lien, Y.-Y.; Tsai, Y.-L. Epidemiology and molecular characterization of chicken anaemia virus from commercial and native chickens in Taiwan. Transbound. Emerg. Dis. 2018, 65, 1493–1501. [Google Scholar] [CrossRef]
- Kim, H.-R.; Kwon, Y.-K.; Bae, Y.-C.; Oem, J.-K.; Lee, O.-S. Molecular characterization of chicken infectious anemia viruses detected from breeder and broiler chickens in South Korea. Poult. Sci. 2010, 89, 2426–2431. [Google Scholar] [CrossRef]
- Van Dong, H.; Tran, G.T.H.; Van Nguyen, G.; Dao, T.D.; Bui, V.N.; Huynh, L.T.M.; Takeda, Y.; Ogawa, H.; Imai, K. Chicken anemia virus in northern Vietnam: Molecular characterization reveals multiple genotypes and evidence of recombination. Virus Genes 2019, 55, 643–653. [Google Scholar] [CrossRef]
- Hoerr, F.J. Clinical Aspects of Immunosuppression in Poultry. Avian Dis. 2010, 54, 2–15. [Google Scholar] [CrossRef]
- Adair, B.M. Immunopathogenesis of chicken anemia virus infection. Dev. Comp. Immunol. 2000, 24, 247–255. [Google Scholar] [CrossRef]
- Hagood, L.T.; Kelly, T.F.; Wright, J.C.; Hoerr, F.J. Evaluation of Chicken Infectious Anemia Virus and Associated Risk Factors with Disease and Production Losses in Broilers. Avian Dis. 2000, 44, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Toro, H.; Gonzalez, O.; Escobar, C.; Cerda, L.; Morales, M.A.; Gonzalez, C. Vertical Induction of the Inclusion Body Hepatitis/Hydropericardium Syndrome with Fowl Adenovirus and Chicken Anemia Virus. Avian Dis. 2001, 45, 215–222. [Google Scholar] [CrossRef]
- Ceruti, R.; Gavazzi, L.; Volorio, A.; Zanella, A. Virus Dell’Anemia Infettiva Aviare: Infezione Subclinica Nel Broiler. In Proceedings of the XLII Convegno Annuale Società Italiana Patologia Aviare (S.I.P.A.), Forlì, Italy, 2–3 October 2003; pp. 67–68. [Google Scholar]
- Li, Y.; Fang, L.; Cui, S.; Fu, J.; Li, X.; Zhang, H.; Cui, Z.; Chang, S.; Shi, W.; Zhao, P. Genomic Characterization of Recent Chicken Anemia Virus Isolates in China. Front. Microbiol. 2017, 8, 401. [Google Scholar] [CrossRef][Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Claessens, J.A.J.; Schrier, C.C.; Mockett, A.P.A.; Jagt, E.H.J.M.; Sondermeijer, P.J.A. Molecular Cloning and Sequence Analysis of the Genome of Chicken Anaemia Agent. J. Gen. Virol. 1991, 72, 2003–2006. [Google Scholar] [CrossRef]
- Brown, K.; Browning, G.; Scott, P.; Crabb, B. Full-length infectious clone of a pathogenic Australian isolate of chicken anaemia virus. Aust. Vet. J. 2000, 78, 637–640. [Google Scholar] [CrossRef]
- Islam, M.R.; Johne, R.; Raue, R.; Todd, D.; Muller, H. Sequence Analysis of the Full-Length Cloned DNA of a Chicken Anaemia Virus (CAV) Strain from Bangladesh: Evidence for Genetic Grouping of CAV Strains Based on the Deduced VP1 Amino Acid Sequences. J. Vet. Med. Ser. B 2002, 49, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Krapež, U.; Barlič-Maganja, D.; Toplak, I.; Hostnik, P.; Rojs, O.Z. Biological and Molecular Characterization of Chicken Anemia Virus Isolates from Slovenia. Avian Dis. 2006, 50, 69–76. [Google Scholar] [CrossRef]
- Van Santen, V.L.; Li, L.; Hoerr, F.J.; Lauerman, L.H. Genetic characterization of chicken anemia virus from commercial broiler chickens in Alabama. Avian Dis. 2001, 45, 373. [Google Scholar] [CrossRef] [PubMed]
- Erfan, A.M.; Selim, A.A.; Naguib, M.M. Characterization of full genome sequences of chicken anemia viruses circulating in Egypt reveals distinct genetic diversity and evidence of recombination. Virus Res. 2018, 251, 78–85. [Google Scholar] [CrossRef]
- Olszewska-Tomczyk, M.; Świętoń, E.; Minta, Z.; Śmietanka, K. Occurrence and Phylogenetic Studies of Chicken Anemia Virus from Polish Broiler Flocks. Avian Dis. 2016, 60, 70–74. [Google Scholar] [CrossRef]
- Huynh, L.T.M.; Van Nguyen, G.; Do, L.D.; Dao, T.D.; Van Le, T.; Vu, N.T.; Cao, P.T.B. Chicken infectious anaemia virus infections in chickens in northern Vietnam: Epidemiological features and genetic characterization of the causative agent. Avian Pathol. 2019, 49, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Tuo, T.; Gao, X.; Han, C.; Yan, N.; Liu, A.; Gao, H.; Gao, Y.; Cui, H.; Liu, C.; et al. Molecular epidemiology of chicken anaemia virus in sick chickens in China from 2014 to 2015. PLoS ONE 2019, 14, e0210696. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Abdel-Mawgod, S.; Adel, A.; Arafa, A.-S.; Hussein, H.A. Full genome sequences of chicken anemia virus demonstrate mutations associated with pathogenicity in two different field isolates in Egypt. Virus Dis. 2018, 29, 333–341. [Google Scholar] [CrossRef]
- Scott, A.N.J.; McNulty, M.S.; Todd, D. Characterisation of a chicken anaemia virus variant population that resists neutralisation with a group-specific monoclonal antibody. Arch. Virol. 2001, 146, 713–728. [Google Scholar] [CrossRef]
- Todd, D.; Scott, A.N.J.; Ball, N.W.; Borghmans, B.J.; Adair, B.M. Molecular Basis of the Attenuation Exhibited by Molecularly Cloned Highly Passaged Chicken Anemia Virus Isolates. J. Virol. 2002, 76, 8472–8474. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.N.J.; Connor, T.J.; Creelan, J.L.; McNulty, M.S.; Todd, D. Antigenicity and pathogenicity characteristics of molecularly cloned chicken anaemia virus isolates obtained after multiple cell culture passages. Arch. Virol. 1999, 144, 1961–1975. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.; Artzi, N.; Shkoda, I.; Lublin, A.; Loeb, E.; Schat, K. The contribution of feathers in the spread of chicken anemia virus. Virus Res. 2008, 132, 152–159. [Google Scholar] [CrossRef]
- Davidson, I. Biotic concerns in generating molecular diagnosis matrixes for 4 avian viruses with emphasis on Marek’s disease virus. J. Virol. Methods 2019, 274, 113708. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, V.L.; Toro, H.; Hoerr, F.J. Biological Characteristics of Chicken Anemia Virus Regenerated from Clinical Specimen by PCR. Avian Dis. 2007, 51, 66–77. [Google Scholar] [CrossRef]
- McNulty, M.S. Chicken anaemia agent: A review. Avian Pathol. 1991, 20, 187–203. [Google Scholar] [CrossRef]
- Brentano, L.; Lazzarin, S.; Bassi, S.; Klein, T.; Schat, K. Detection of chicken anemia virus in the gonads and in the progeny of broiler breeder hens with high neutralizing antibody titers. Vet. Microbiol. 2005, 105, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Creelan, J.L.; Connor, T.J.; Ball, N.W.; Scott, A.N.; Meehan, B.M.; McKenna, G.F.; McNulty, M. Investigation of the unstable attenuation exhibited by a chicken anaemia virus isolate. Avian Pathol. 2003, 32, 375–382. [Google Scholar] [CrossRef]
- Vaziry, A.; Silim, A.; Bleau, C.; Frenette, D.; Lamontagne, L. Chicken infectious anaemia vaccinal strain persists in the spleen and thymus of young chicks and induces thymic lymphoid cell disorders. Avian Pathol. 2011, 40, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, M.; Catelli, E.; Lupini, C.; Ricchizzi, E.; Prosperi, S.; Naylor, C. Reversion to virulence of a subtype B avian metapneumovirus vaccine: Is it time for regulators to require availability of vaccine progenitors? Vaccine 2014, 32, 4660–4664. [Google Scholar] [CrossRef]
- Franzo, G.; Naylor, C.J.; Lupini, C.; Drigo, M.; Catelli, E.; Listorti, V.; Pesente, P.; Giovanardi, D.; Morandini, E.; Cecchinato, M. Continued use of IBV 793B vaccine needs reassessment after its withdrawal led to the genotype’s disappearance. Vaccine 2014, 32, 6765–6767. [Google Scholar] [CrossRef]
- Groves, P.J.; Williamson, S.L.; Sharpe, S.M.; Gerber, P.F.; Gao, Y.K.; Hirn, T.J.; Walkden-Brown, S.W. Uptake and spread of infectious laryngotracheitis vaccine virus within meat chicken flocks following drinking water vaccination. Vaccine 2019, 37, 5035–5043. [Google Scholar] [CrossRef]
- Falchieri, M.; Lupini, C.; Cecchinato, M.; Catelli, E.; Kontolaimou, M.; Naylor, C.J. Avian metapneumoviruses expressing Infectious Bronchitis virus genes are stable and induce protection. Vaccine 2013, 31, 2565–2571. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Cortes, A.L.; Faiz, N.; Villalobos, T.; Badillo, H.; Barbosa, T. Efficacy of Various HVT Vaccines (Conventional and Recombinant) Against Marek’s Disease in Broiler Chickens: Effect of Dose and Age of Vaccination. Avian Dis. 2016, 60, 662–668. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Liu, Y.-C.; Hsu, Y.-C.; Chang, P.-C.; Hsieh, M.-K.; Shien, J.-H.; Ou, S.-C. Preparation of Chicken Anemia Virus (CAV) Virus-Like Particles and Chicken Interleukin-12 for Vaccine Development Using a Baculovirus Expression System. Pathogens 2019, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Gelb, J.; Jackwood, D.J.; Brannick, E.M.; Ladman, B.S. Efficacy of Recombinant HVT-IBD Vaccines Administered to Broiler Chicks from a Single Breeder Flock at 30 and 60 Weeks of Age. Avian Dis. 2016, 60, 603–612. [Google Scholar] [CrossRef]
- Felice, V.; Lupini, C.; Mescolini, G.; Silveira, F.; Guerrini, A.; Catelli, E.; Di Francesco, A. Molecular detection and characterization of Mycoplasma gallisepticum and Mycoplasma synoviae strains in backyard poultry in Italy. Poult. Sci. 2020, 99, 719–724. [Google Scholar] [CrossRef]
- Mescolini, G.; Lupini, C.; Felice, V.; Guerrini, A.; Silveira, F.; Cecchinato, M.; Catelli, E. Molecular characterization of the meq gene of Marek’s disease viruses detected in unvaccinated backyard chickens reveals the circulation of low- and high-virulence strains. Poult. Sci. 2019, 98, 3130–3137. [Google Scholar] [CrossRef] [PubMed]
- Haesendonck, R.; Verlinden, M.; Devos, G.; Michiels, T.; Butaye, P.; Haesebrouck, F.; Pasmans, F.; Martel, A. High Seroprevalence of Respiratory Pathogens in Hobby Poultry. Avian Dis. 2014, 58, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Derksen, T.; Lampron, R.; Hauck, R.; Pitesky, M.; Gallardo, R.A. Biosecurity Assessment and Seroprevalence of Respiratory Diseases in Backyard Poultry Flocks Located Close to and Far from Commercial Premises. Avian Dis. 2018, 62, 1–5. [Google Scholar] [CrossRef]
- Gimeno, I.M.; Schat, K.A. Virus-Induced Immunosuppression in Chickens. Avian Dis. 2018, 62, 272–285. [Google Scholar] [CrossRef] [PubMed]


| Production Type | Sample ID | Sample | Farm | Flock | Italian Region | Poultry Company | Age of Birds/Time of Sampling | CIAV PCR Results (Genogroup) |
|---|---|---|---|---|---|---|---|---|
| Broiler | 909-06/18 ** | Feathers * | 1 | A | Veneto | 1 | 40 days | Positive (IIIa) |
| 986-1/18 | Dust | 2 | A | Friuli-Venezia Giulia | 2 | End of production cycle (post-cleaning) | Positive (II) | |
| 986-2/18 ** | B | Positive (II) | ||||||
| 986-6/18 | C | Positive (II) | ||||||
| 1018/18 | Dust | 3 | A | Friuli-Venezia Giulia | 2 | End of production cycle (post-cleaning) | Negative | |
| 1019/18 | B | Negative | ||||||
| 1155/19 ** | Dust | 4 | A | Friuli-Venezia Giulia | 2 | End of production cycle (before-cleaning) | Positive (IIIb) | |
| 1157/19 ** | Feathers | B | Positive (IIIa) | |||||
| 1178/19 | Dust | 5 | A | Friuli-Venezia Giulia | 2 | N.A. | Negative | |
| 1179/19 | B | Positive (II) | ||||||
| 1180/19 ** | C | Positive (IIIb) | ||||||
| 1185/19 | Dust | 6 | A | Friuli-Venezia Giulia | 2 | N.A. | Positive (IIIa) | |
| 1186/19 ** | B | Positive (IIIa) | ||||||
| 1187/19 | C | Positive (II) | ||||||
| 1188/19 ** | Dust | 7 | A | Friuli-Venezia Giulia | 2 | N.A. | Positive (IIIa) | |
| 1189/19 | B | Positive (IIIa) | ||||||
| 1191/19 | Dust | 8 | 3,4,5,6 | Emilia-Romagna | 2 | End of production cycle (before-cleaning) | Positive (II) | |
| 1192/19 | 7,8,9 | Positive (IIIa) | ||||||
| 1194/19 | Dust | 9 | / | Emilia-Romagna | 2 | 35 days | Negative | |
| 1195/19 | Dust | 10 | / | Emilia-Romagna | 2 | 47 days | Positive (IIIb) | |
| 1196/19 ** | Spleen * | 11 | / | Emilia-Romagna | 2 | N.A. | Positive (IIIa) | |
| 1201/19 | Dust | 12 | / | Friuli-Venezia Giulia | 2 | 42 days | Positive (IIIa) | |
| 1099/19 ** | Dust | 13 | A | Friuli-Venezia Giulia | 3 | End of production cycle (post-cleaning) | Positive (II) | |
| 1100/19 | B | End of production cycle (post-cleaning) | Positive (II) | |||||
| 1101/19 | C | End of production cycle (before-cleaning) | Positive (II) | |||||
| Pullet Broiler Breeders | 989/18 | Dust | 14 | A | Abruzzo | 4 | Day 0 | Negative |
| 1014-1/18 | Dust | B | 12 weeks | Positive (IIIa) | ||||
| 1152-1/19 | Feathers * | C | 16 weeks | Negative | ||||
| 1153-1/19 | Dust | 16 weeks | Positive (IIIb) | |||||
| 1218/19 | Dust | End of production cycle (post-cleaning) | Negative | |||||
| 1152-2/19 | Feathers * | D | 16 weeks | Negative | ||||
| 1153-2/19 ** | Dust | 16 weeks | Positive (IIIa) | |||||
| 1219/19 | Dust | End of production cycle (post-cleaning) | Negative | |||||
| 1220/19 | Dust | E | End of production cycle (post-cleaning) | Negative | ||||
| 1221/19 | Dust | F | End of production cycle (post-cleaning) | Negative | ||||
| 1015-2/18 | Feathers * | 15 | / | Abruzzo | 4 | 22 weeks | Negative | |
| Backyard Chickens | 801/17 | Feathers * | / | G | Sicily | / | 3.5 to 4 months | Negative |
| 810/17 | Feathers * | / | H | Sicily | / | 3 to 4.5 months | Negative | |
| 847/17 | Feathers * | / | I | Lombardy | / | 12 months | Negative | |
| 850/17 | Feathers * | / | L | Tuscany | / | 6 months | Negative | |
| 852/17 | Feathers * | / | M | Campania | / | 6 to 9 months | Negative | |
| 853/17 | Feathers * | / | N | Lombardy | / | 4 to 7 months | Negative | |
| 854/17 | Feathers * | / | O | Trentino-Alto Adige | / | 9 to 24 months | Negative | |
| 855/17 ** | Feathers * | / | P | Tuscany | / | 8 to 12 months | Positive (II) |
| PCR | PCR Primer Sequence 5′-3′ | Position in the Genome | bp |
|---|---|---|---|
| 1 | 5′-CAGGGTAAGCGAGCTAAAAG-3′ | 751–770 | 1528 |
| 3′-GCTGCGTTTATTGAGTGC-5′ | 2262–2279 | ||
| 2 | 5′-GGTACGTATAGTGTGAGGC-3′ | 996–1014 | 790 |
| 3′-GCTGTGAGTGTTGCAAAGCT-5′ | 1767–1786 |
| CIAV Strain | Country | GenBank Accession No. | Genogroup | Reference |
|---|---|---|---|---|
| Del-Ros | USA | AF313470 | IIIb | N.A. |
| 26P4 | USA | D10068 | IIIb | [29] |
| Nobilis P4 vaccine | The Netherlands | AJ890284 | IIIb | [18] |
| Cux-1 | Germany | M55918 | IIIb | [3] |
| Cuxhaven 1 | Germany | M81223 | IIIb | [4] |
| CAU269-7 | Australia | AF227982 | I | [30] |
| 3711 | Australia | EF683159 | I | N.A. |
| BD-3 | Bangladesh | AF395114 | IIIa | [31] |
| CAV/Ibadan.NIE/11.02/100 | Nigeria | AJ888519 | II | [18] |
| CAV/Lanlate.NIE/11.02/71 | Nigeria | AJ888528 | IIIa | [18] |
| 69 | Slovenia | DQ016140 | IIIa | [32] |
| 98D02152 | USA | AF311892 | IIIb | [33] |
| Isolate 4 | Taiwan | KJ728816 | IIIb | N.A. |
| Isolate 6 | Taiwan | KJ728817 | IIIb | N.A. |
| Isolate 8 | Taiwan | KJ728819 | IIIa | N.A. |
| Isolate 9 | Taiwan | KJ728820 | IIIa | N.A. |
| CAV-EG-2 | Egypt | MH001553 | IIIb | [34] |
| CAV-EG-11 | Egypt | MH001559 | IIIb | [34] |
| CAV-EG-13 | Egypt | MH001560 | IIIb | [34] |
| CAV-EG-14 | Egypt | MH001565 | II | [34] |
| CAV-EG-26 | Egypt | MH001564 | IIIa | [34] |
| CAV/LOD5/13 | Poland | KM458172 | IIIa | [35] |
| CAV/MPL2/13 | Poland | KM458175 | IIIa | [35] |
| CAV/OPL3/13 | Poland | KM458178 | IIIa | [35] |
| CAV/SLA3/13 | Poland | KM458182 | IIIa | [35] |
| G17.33.3 | Vietnam | MH536104 | IIIa | [36] |
| HB1517 | China | KU645516 | IIIa | N.A. |
| AH6 | China | DQ124935 | IIIa | N.A. |
| HLJ15108 | China | KY486137 | IIIa | [37] |
| LF4 | China | AY839944 | IIIb | N.A. |
| SD1514 | China | KU645521 | IIIa | N.A. |
| 704 | Australia | U65414 | II | N.A. |
| 98D06073 | USA | AF311900 | II | [33] |
| 1102PT01 | Taiwan | KY888892 | IV | [19] |
| 1103TN02 | Taiwan | KY888894 | IV | [19] |
| SD22 | China | DQ141673 | IV | N.A. |
| SD24 | China | AY999018 | IV | N.A. |
| CIAV Strains | Amino Acid Position | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 75 | 97 | 125 | 139 | 144 | 157 | 287 | 290 | 370 | 394 | 413 | 447 | |
| Del-Ros | H | V | M | I | K | E | V | S | A | G | Q | S | G |
| 26P4 | . | . | . | . | . | . | M | T | . | S | . | A | T |
| CIAV/IT/CK/855/17 | . | I | L | . | Q | Q | . | T | . | S | . | A | S |
| CIAV/IT/CK/909-06/18 | . | I | L | . | Q | Q | . | A | . | T | . | A | S |
| CIAV/IT/CK/986-1/18 | - | I | L | L | Q | Q | . | T | P | - | - | - | - |
| CIAV/IT/CK/986-2/18 | . | I | L | . | Q | Q | . | T | P | T | . | A | S |
| CIAV/IT/CK/986-6/18 | - | I | L | L | Q | Q | . | T | P | - | - | - | - |
| CIAV/IT/CK/1014-1/18 | N | I | L | . | Q | Q | . | A | . | T | . | A | S |
| CIAV/IT/CK/1099/19 | . | I | L | . | Q | Q | . | T | . | S | . | A | S |
| CIAV/IT/CK/1100/19 | . | I | L | . | Q | Q | . | T | . | - | - | - | - |
| CIAV/IT/CK/1101/19 | - | I | L | . | Q | Q | . | T | . | S | . | A | S |
| CIAV/IT/CK/1153-1/19 | - | . | . | . | . | . | M | T | . | - | - | - | - |
| CIAV/IT/CK/1153-2/19 | N | I | L | . | Q | Q | . | A | . | S | . | A | S |
| CIAV/IT/CK/1155/19 | Q | . | . | . | Q | Q | . | . | . | R | . | . | S |
| CIAV/IT/CK/1157/19 | N | I | L | . | Q | Q | . | A | . | T | . | A | S |
| CIAV/IT/CK/1179/19 | - | I | L | . | Q | Q | . | T | . | - | - | - | - |
| CIAV/IT/CK/1180/19 | Q | . | . | . | . | . | . | . | . | . | . | . | S |
| CIAV/IT/CK/1185/19 | - | I | L | . | Q | Q | . | A | . | S | . | A | S |
| CIAV/IT/CK/1186/19 | . | I | L | . | Q | Q | . | A | . | T | . | A | S |
| CIAV/IT/CK/1187/19 | - | I | L | . | Q | Q | . | T | . | S | . | A | S |
| CIAV/IT/CK/1188/19 | N | I | L | . | Q | Q | . | A | . | S | . | A | S |
| CIAV/IT/CK/1189/19 | - | I | L | . | Q | Q | . | A | . | - | - | - | - |
| CIAV/IT/CK/1191/19 | - | I | L | . | Q | Q | . | T | P | S | . | A | S |
| CIAV/IT/CK/1192/19 | - | I | L | . | Q | Q | . | A | . | - | - | - | - |
| CIAV/IT/CK/1195/19 | - | . | L | . | Q | Q | . | . | . | - | - | - | - |
| CIAV/IT/CK/1196/19 | . | I | L | . | Q | Q | . | A | . | T | . | A | S |
| CIAV/IT/CK/1201/19 | - | I | L | . | Q | Q | . | A | . | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quaglia, G.; Mescolini, G.; Catelli, E.; Berto, G.; Muccioli, F.; Lupini, C. Genetic Heterogeneity among Chicken Infectious Anemia Viruses Detected in Italian Fowl. Animals 2021, 11, 944. https://doi.org/10.3390/ani11040944
Quaglia G, Mescolini G, Catelli E, Berto G, Muccioli F, Lupini C. Genetic Heterogeneity among Chicken Infectious Anemia Viruses Detected in Italian Fowl. Animals. 2021; 11(4):944. https://doi.org/10.3390/ani11040944
Chicago/Turabian StyleQuaglia, Giulia, Giulia Mescolini, Elena Catelli, Giacomo Berto, Filippo Muccioli, and Caterina Lupini. 2021. "Genetic Heterogeneity among Chicken Infectious Anemia Viruses Detected in Italian Fowl" Animals 11, no. 4: 944. https://doi.org/10.3390/ani11040944
APA StyleQuaglia, G., Mescolini, G., Catelli, E., Berto, G., Muccioli, F., & Lupini, C. (2021). Genetic Heterogeneity among Chicken Infectious Anemia Viruses Detected in Italian Fowl. Animals, 11(4), 944. https://doi.org/10.3390/ani11040944





