1. Introduction
The prospects of complementing already established endangered breeds or populations into a multi-breed composite population was proposed by Shrestha [
1]. This author argued that, even though the development of a composite population does not directly lead to the conservation of an endangered breed, it may promote ‘conservation by utilisation’, through the preservation of the inherent potential of foundational breeds.
The aforementioned concept lays the basis for breeding programs either it is conservation plans seeking the sustainable management of native breeds, or the commercial development and utilization of specific crosses for practical purposes. Particular features of each population contribute to the genetic variability of the species to which it belongs. However, these particular features could dilute due to sub structuring, intermixing and/or consequent genetic drift over time [
2].
The origins of the Há horse breed [
3] date back to the year 742, and can be set in the cavalry of Balŷ ibn Bišr al-Qušayri, eighteenth governor (valí) of al-Ándalus (741–742), who brought Arabian horses along with his 10,000 soldiers to the Iberian peninsula when Berber insurrection extended to al-Ándalus [
4]. As a result of this introduction, even though a small number of pure Arabian horses (PRá) were brought by the army of Balŷ, they were sufficient to originate a new type of horse: the Iberian Arabian horse. These PRá horses from Spain crossed with the primitive Spanish horse, found in the peninsula, originated the predecessors of the current Há horse [
4].
Since its origins, Há horse breeders have sought complementarity between PRE and PRá horses [
5]. As suggested by some authors [
5], PRE horses genetically provided an optimal endocrine functionality and regulatory ability of the muscular physiology, together with an enhanced reproductive efficiency and adaptability to harsh environments, which promoted the expansion of the breed to the New World with Cristopher Columbus in 1493. Simultaneously, PRá horses conferred their strength, dexterity, and their cognitive superiority [
6] to the crossing.
Both ancestor breeds have been reported to be more closely related to each other than other affine breeds such as Anglo-Arabian or The English Trotting Horse, as suggested by the genic and genotypic frequencies for blood systems “A” and “O” [
7,
8,
9].
The improved features derived from composite vigour (heterosis) prompted authors such as Ibn al-’Awwam, agriculturist of the later 12th century, to refer to the cross between PRE and PRá as “the best” par excellence in his Kitāb al-filāha [
10]. This manuscript is the most comprehensive agricultural treatise in Arabic and gathers all the knowledge of its time in respect to agriculture, horticulture and animal husbandry [
11]. Contextually, some authors have hypothesized that Há may have been an attempt to emulate other relevant Arabian breeds of the Middle East [
12].
At first sight, Há horses may have a smaller size, a remarkable trend to fit eumetric proportions and rather lively gaits than its ancestor breeds (PRE and PRá horses). There are some archive records reporting the use of earlier Há breeding studs, as far back as 1778. However, the formal creation of the breed dates to the 1883, when an official breeding program was implemented coinciding with the introduction of Arabian horses to upgrade and expand other horse breeding programs. Although the breed is linked to the Andalusian countryside, where it gained popularity as a working cattle horse, its expansion and consolidation as a breed has been slow [
13]. The lack of quality of stock in some of the earlier breeding lines and the popularity of the cross between Há and thoroughbred (part-bred) horses to conform the Tres Sangres (Three bloods) composite breed can be found among the main causes for this slow consolidation process.
The Há breed was formerly very common in the south of Spain (Andalusian), area in which it was presumably created and from which the breed would spread even internationally. However, an increased national and international demand for the PRE horses led to the decrease in Há breed effective numbers, until it became a minority rare breed by the mid-1980s. This situation led to the constitution of the Há horse breed studbook in 1986 and its later official recognition within the category of special protection through the inclusion of the breed in the Official Catalogue of Spanish Cattle Breeds (Orden APA/2129/2008) [
14]. The efforts of Cría Caballar (Military stables), a division of the Spanish Ministry of Defence, started in 1990 with a number of strategies which aimed to conserve and aid the recovery and breeding of the Há breed on military stud centers. As a result, in the middle of the 19th century the breed started to be considered standardized, due to the role it played in military campaigns. The current breed standard was published in 2002, and was modified in 2005. Since 2008, the studbook has been held by the Spanish Union of Purebred Hispano-Arabian Horses Breeders (UEGHá).
Contextually, the physical endurance, athletic qualities, balance in character of the PRá horse were, have been and still are combined with the versatility, predisposition to work and movement precision of the PRE horse. The evolution of the breeding strategies sought the recovery of the numbers of Há horses and can be evaluated in five stages. The first stage ranged from 1992 to 1995 and was marked by the use of 16 foundation Há stallions bred with (F1) 50:50 mares, resulting from the crossing between PRE dams and PRá sires. Afterwards, during the second stage which ran from 1996 to 1999, five selected PRá stallions were used to cover Há mares and one PRE stallion was selected to cover PRá mares. The third stage (2000–2005) was characterized by the predominant use of PRE horses covering more than 80% of the Há mares. A parallel breeding program dedicated to producing F1 Há individuals from PRE mares covered by both PRá stallions and five Há stallions of 50% and 75% PRá blood ratios obtained at the aforementioned stages. Simultaneously, the military covered PRá mares with PRE stallions, which had not been a frequent practice at the previous stages. Additionally, alongside these controlled breeding strategies, some mares were freely covered by Há stallions to produce F2 and F3 individuals optimally fitting the breed standard with the aim to improve the breed.
The objective of the fourth stage, which run from 2006 until our days, was to include the F2 individuals with PRá blood percentages ranging between 62.5% and 37.5%. Such individuals used to be difficult to find at previous stages, but are the natural progression of outbreeding 25% and 75% Há horses to PRE or PRá horses. Contextually, Há stallions and mares of 25% and 75% PRá blood bred to each other produce a 50:50 fixed Há genotype. This strategy sought to ensure that the products derived from the aforementioned breeding program effectively ascribed to the breed standard. At the age of 3, individuals start to be trained in the development of the skills comprised by the disciplines of Spanish cowboy/western dressage (Doma vaquera) and cattle driving, for which it stands out above other breeds (such as its ancestor breeds, PRE and PRá horses) [
12].
Although the evaluation of genetic diversity and demographic parameters raised research interest in the past [
15], previous analyses did not consider the repercussions of each of the two ancestor breed. Hence, parameters may have potentially been misestimated as a result of the genealogical information present in pedigrees being largely incomplete [
16,
17].
Therefore, the aim of this study is the development of a model to perform the discriminant canonical analysis of the contributions of PRE and PRá horses to the historical and present genetic diversity and demographic structure of the Há horse breed. Pedigree completeness was evaluated downwards, checking the repercussions of ancestors and founders, evaluating the structure of the population, its genetic variability, and connections between its genetic and demographic parameters, measuring the existing gene flow, and quantifying the risk of genetic diversity loss. The endangerment risk that the Há horse breed faces was evaluated to suggest effective conservation strategies, which in the case of the Há composite breed may reinforce its entity within the horse breed international panorama.
4. Discussion
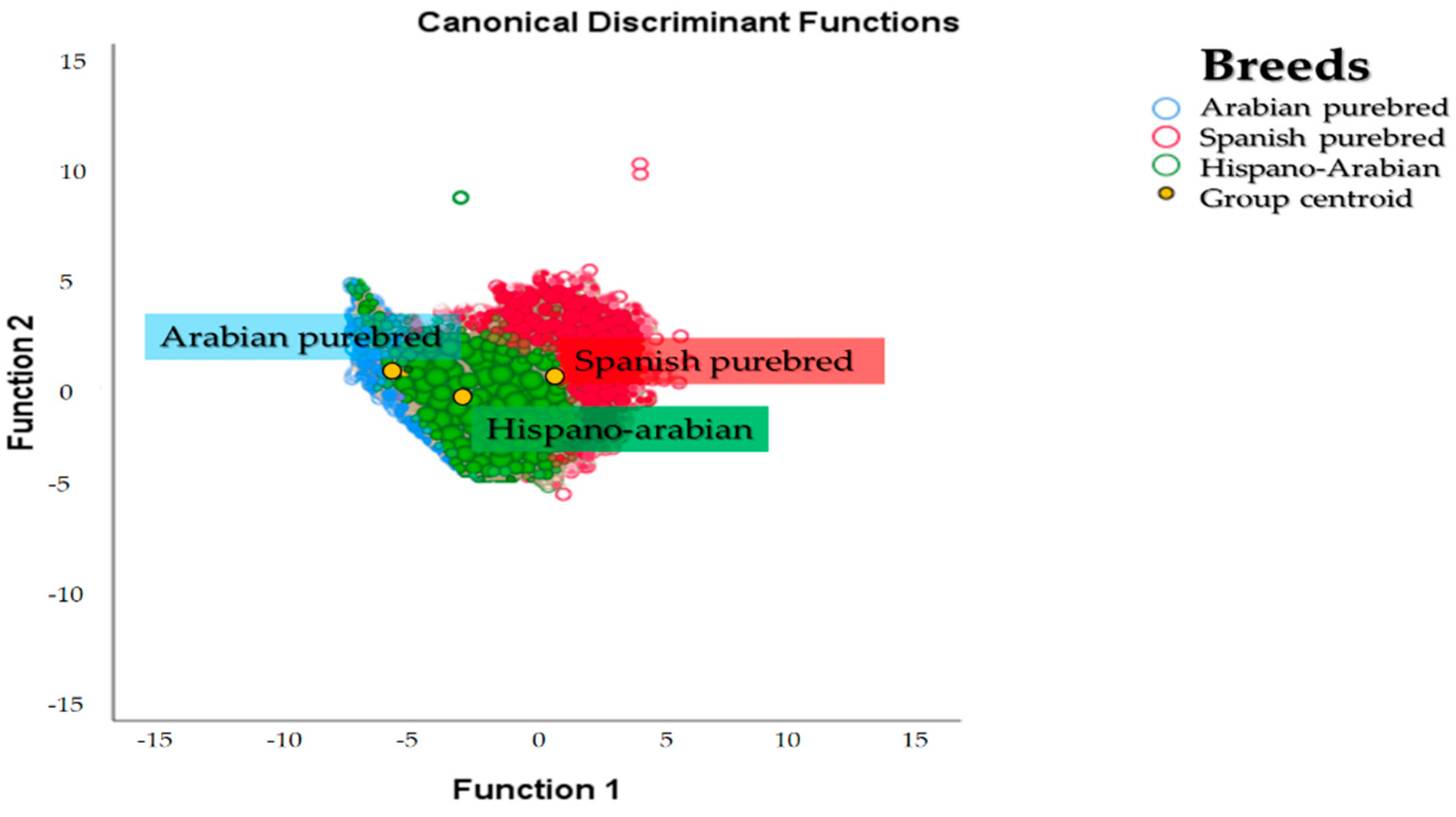
The Há horse breed is a composite breed which derives from the cross between PRE and PRá horse individuals. To better understand Há horses, we may have to go back and study its ancestor breeds. This may enable the gathering of a very comprehensive amount of information to study the evolution of genetic diversity and of factors affecting it. The examination of the dendrogram constructed from Nei genetic distances and the spatial distribution reported after the discriminant analyses (
Figure 3) suggest the existence of clear population structure, which may still be partially supported by its two ancestor breeds. In this context, a higher repercussion of PRá horses on Há and hence proximity between the breeds is evidenced when PRá is compared to PRE horses (
Figure 3 and
Figure 5).
The Mediterranean breeds considered in the study (PRá, PRE, and Há horse breeds) present strong differential physiological, behavioural and morphological differences [
5,
6,
10,
12,
65] which are closely related to the geographical locations in which they locate and have developed along the course of History. This distinction and the closest link between PRá and Há horses was also supported at a genetic level by Pablo Gómez, et al. [
66]. This close connection between North African horses and those inhabiting in the Southern territories of the Iberian Peninsula has been reported to have occurred since prehistorical times and before the commercial routes established by the Greek and the Phoenician colonies took place.
According to Aparicio Sánchez [
67], such former connection was reinforced for centuries when, once commercial routes had been established, a significant horse exchange existed between North Africa and the Iberian Peninsula and extended at least from the arrival of the Romans in Iberia in 219/8 BC for the conquest of Hispania and up to the expulsion of the Moriscos, as decreed by King Philip III of Spain on 9 April 1609 [
68]. In line with this historical context, even if the breeding practices seeking the obtention of Há horses only became more relevant from 1800 on, the present study evidences that the greater contribution of PRás to the development of Há breed and its genetic diversity may not only be patent from the very first crosses carried out after the first PRá horses came to the Iberian Peninsula in 742, but it may also continue in our times.
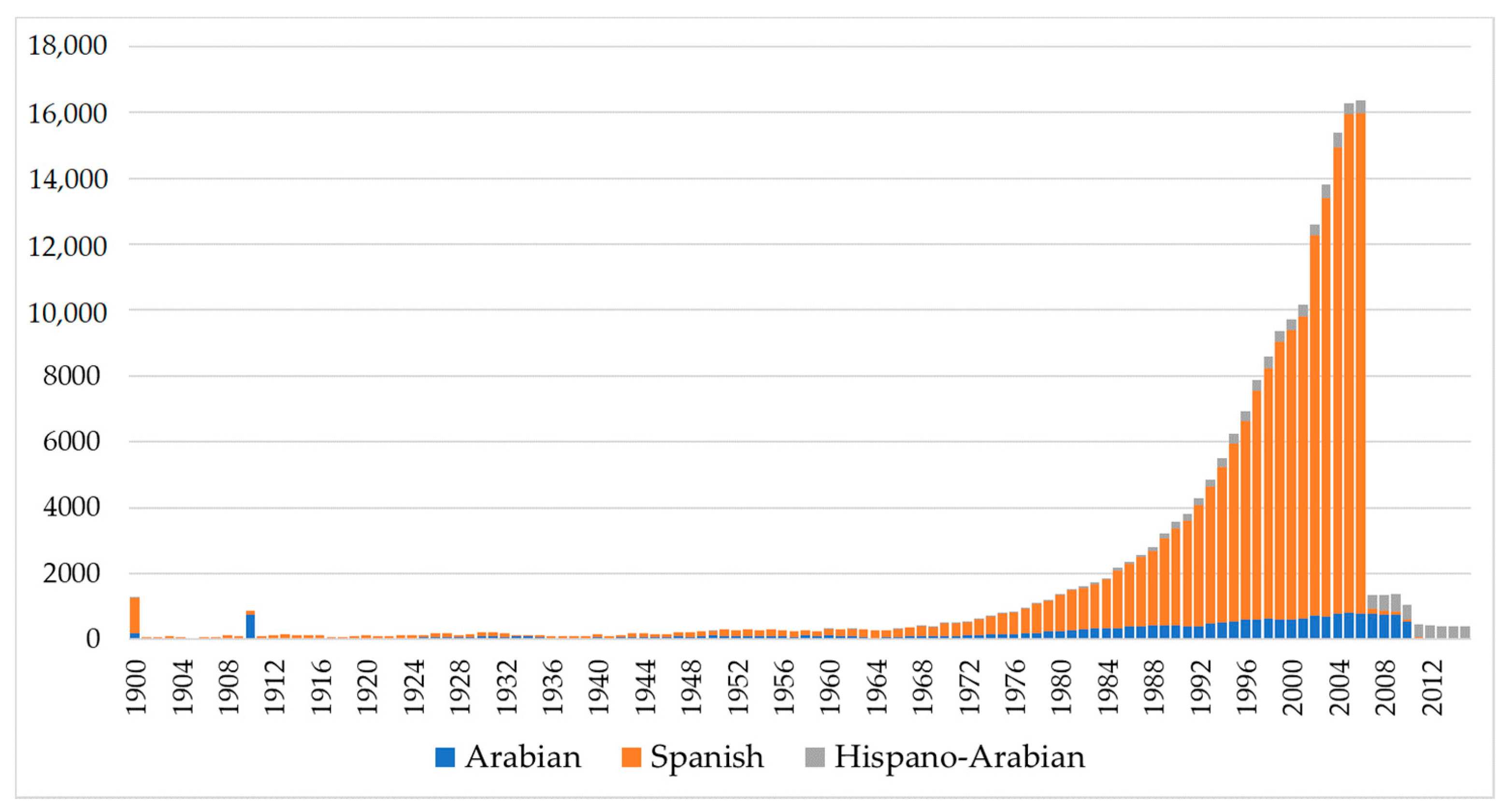
The drastic reduction in the number of PRE horses (
Figure 1) was a direct consequence of the economic crisis whose effects in the equine sector became patent in 2008, when the real estate bubble burst [
60]. According to Palomo [
60], prior to 2008, PRE horses had been acquired as a luxury item by real estate developers or companies, who had no choice but to missell them when the economic crisis arose. Abandonment, giveaway prices or slaughterhouse became the destination of thousands of individuals, which also brought about the drastic stop of breeding practices. It was only from 2012 on that it began to rebound due to leisure or sport. At the worst of times, individuals were sold for 150 €, while the average price ranges between 3000 and 5000 €. Há horses did not suffer from the effects of the crisis as drastically as PRE horses as these were usually kept on large farms, sometimes as a secondary production alongside cow farming, hence they were sold more easily and at a better price given their versatility [
61].
Geographical barriers or socio-political considerations may not have played an important role in the loss of diversity of the population given the low levels of genetic diversity loss found in our study, which could be ascribed to the process of genetic drift within the population. Overall, the generation interval in Há is rather long, with a global mean for the four selection paths of around 14.81 years. Nevertheless, this is in line with other studies considering the PRE horse breed (10.1 years) [
69], largely coinciding with the values for PRE horses in our study, with generation intervals of 10.58 years in the current population. Likewise, the generation interval in PRá horse breed was 13.03 years, which was slightly higher than the values reported in literature [
70]. This suggest that the unbalanced contribution of PRá horses to the development of Há horses, may also contribute to the fact that breeding practices and possibly reproductive physiology [
71] may resemble those in the PRá horse more than those in the PRE breed, which may explain the slightly longed generation intervals.
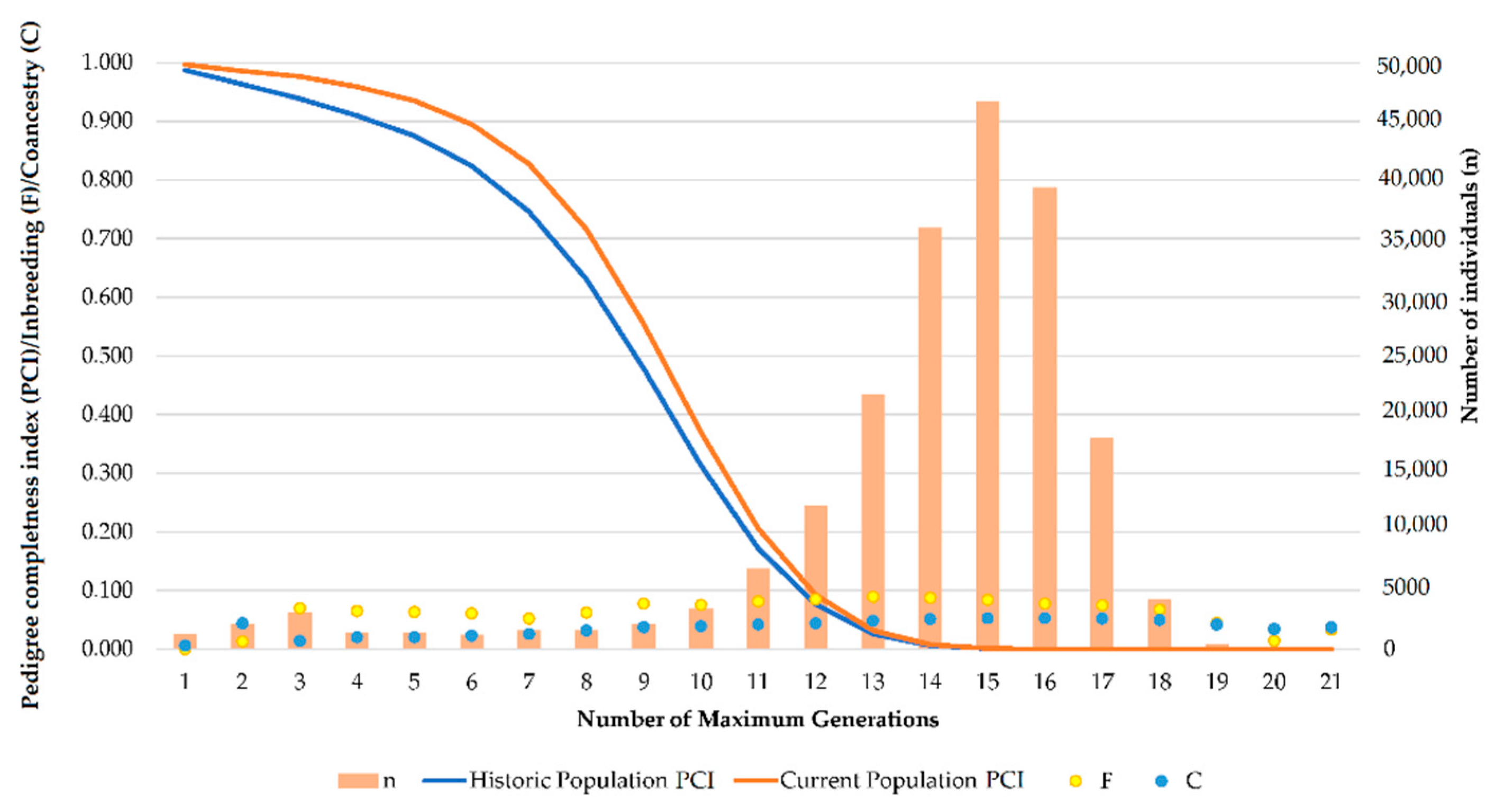
PCI in the three breeds studied are very high for the first five generations (from parents to great-great grandparents), which is common to autochthonous horse breeds as supported by Giontella, et al. [
72]. The high quality of the pedigree used in this study, provided its length and depth, enables an accurate calculation of genetic diversity parameter. The almost constant levels of inbreeding and coancestry reported until the 21st generation suggest that the analysis of genetic diversity and population structure analysis is accurate and valid and accurate conclusions can be drawn. As demonstrated by Duru [
73], all parameters describing the probability of gene origin of a certain population are affected by pedigree depth. In fact, a suitable estimation of genetic variability widely depends on available and accessible pedigree information measured by pedigree completeness.
The use of robust pedigrees with completeness indices of around 80% have reported to allow reliable estimations of inbreeding values, resulting in medium to high correlations with genomic inbreeding in horse breeds [
74]. Average values for PCI in PRá, PRE, and Há historical and current populations were 77.07%, 90.13%, 96.13%, 99.01%, 73.26%, and 74.71%, respectively. Todd, et al. [
75] suggested that among the causes supporting the occurrence of these high correlations, a large proportion of the inbreeding coefficient in horse current populations may be accounted for by ancestors many generations back in the pedigree. Inbreeding to distant ancestors results in shorter runs of homozygosity regions which might not be captured unless very high densities of SNP are used. These authors [
75] suggested that pedigree analyses can report inferences of inbreeding that are comparably accurate to those reported by SNPs analyses, when complete pedigrees large populations considerably exceed the number of genotyped individuals. Such event occurs as pedigree data may enable to make inferences for deceased individuals (such as the founders of the population), whose molecular DNA material cannot be recovered to perform genotyping. Furthermore, these complete pedigrees may allow to evaluate the trends described by genetic diversity alongside the history of specific populations, offering the opportunity to determine the effects of particular individuals over time on the fitness of their descendants.
The use of breeds with dissimilar performance characteristics enhances the opportunities for the maximization of their average genetic merit in the resulting product. This is taken advantage to meet requirements for specific functionality, production and/or marketing situations. When breeds used in the foundation of a composite breed do not contribute equally, as suggested by our results, since there is a loss of heterozygosity between the first and second generations [
76]. Then, if inbreeding is avoided, further loss of heterozygosity in mated populations does not occur. This theoretical framework is well described by the populations evaluated in the present study. For instance, although mean inbreeding levels in the historical population of the Há and of its ancestor breeds, PRá and PRE horse breeds were 2.85%, 6.79%, and 8.42%, respectively, there has been a slight increase in inbreeding levels in the current populations of the same breeds to reach the levels of the 2.89%, 8.44%, and 8.55%, respectively.
Although genetic erosion may have occurred at a very low rate, the population of Há horses has remained stable. These results suggest, timely action must be taken to control the increase in inbreeding through the years in the two ancestor breeds as levels are starting to approach compromising levels, which may result in the expression of deleterious effects derived from inbreeding in the population. One of the most relevant effects resulting from reduced genetic diversity is inbreeding depression, which in turn, may end up compromising the performance of domestic animals [
77]. Santana Jr, et al. [
77] reported that the maximum level of inbreeding that could be absorbed by animals before detrimental effects begin to negatively affect performance is around 20%. However, deleterious effects related to diseases, reproduction or cognitive function may be patent when inbreeding levels are around 12.5% or above [
78,
79,
80,
81].
In this context, the large number of stallions used in reproduction may have contributed to the stabilization of inbreeding. Still, the presence of highly inbred animals in the population in percentages of 4.60%, 19.41%, and 30.55% in the currently living Há, PRE, and PRá horse populations implies the excessive use of certain individuals is still patent. These findings support the fact that currently, the major concern in managing the genetic diversity of the Há horse breed is the short-term decrease in genetic variability due to the loss of genetic contributions from founders and ancestors, more than the long-term effect of inbreeding itself as it was suggested in literature [
82,
83,
84,
85,
86,
87,
88].
When aiming to develop a genetically healthy breeding strategy, ΔR is a very useful parameter since it allows breeders to preserve the genetic pool of a certain population [
72,
89]. Because of that, if the stallions and mares presenting the lowest levels of ΔR are mated (in our case PRá with ΔR 1.42% and Há horse breed with ΔR 4.25%), the inbreeding levels in their future offspring may be reduced, which may act balancing the gene contributions of the founders in the population, and consequently the genetic diversity.
Levels of genetic diversity in the present population are over 93% for the three breeds. However, in the context of Há horses as a composite breed whose studbook is still open, these high levels may not ensure the preservation of the gene pool of the founding population, as this population may constantly be changing [
90]. In such cases, the genetic conservation Index (GCI) can help to determine the contribution of founders of each breed to the composite population. McManus, et al. [
91] described that GCI computes the genetic contributions of all the identified founders; for this reason, it has been assumed that the animals, which, get higher values of GCI, also gather wider fractions of the gene pool of the founding population. In our case, the higher GCI value was for PRá horse breed (11.50 ± 3.76), (9.74 ± 1.28 for Spanish horse breed) and (9.38 ± 5.88 for Há horse breed), which supports the higher contribution to the founding gene pool of PRá horses than PRE horses to the conformation and development of the Há horse breed, as it has been suggested by our results and other authors [
66].
The determination of Mendelian sampling of the non-founders of a population has recently been reported to be of help when determining partial inbreeding in equine populations [
92]. Inbreeding can be broken down into the sources of the coancestry between the parents of each individual, which becomes even more relevant in the case of composite breeds whose ancestors may belong to differentiated breeds. In our study, the number of non- founder (
Nef) for PRá, PRE, and Há horse breed were 10, 11.40, and 23.57, respectively. These values support those reported in the literature [
26,
93], even more so when these values are evaluated in the context of the inbreeding levels presented for the three horse breeds discussed. In line with these results, we may determine that relative founder contributions may tend to stabilize after a short number of generations, which has been described in Thoroughbred horses [
94]. Either the population is closed or remains open, as is the case of the Há horse breed [
17,
92].
FAO/UNEP [
95] proposed the general rule of maintaining rate of inbreeding per generation should not exceed 1–3% [
96]. Higher rates fix deleterious recessive genes too rapidly for selection to eliminate them, and the vigour and fertility of the populations decrease. When inbreeding rate is below 1% populations have been partially purged of deleterious genes and tolerate higher rates of inbreeding, hence, animal breeders can safely ignore some inbreeding and random loss of genes. In endangered populations conservationists develop a rather conservative approach. The rate of loss per generation of heterozygosity due to inbreeding as measured by F is equal to 1/(2
Ne), where
Ne is the effective population size.
The definition of
Ne is complex, but certain criterion must be considered to permit a correct interpretation of this parameter. For instance, the sex ratio must be equal and individuals must randomly mate. A number of additional “ideal” characteristics could be stated. In practice,
Ne is always smaller than the actual number of breeding individuals. Thus,
Ne must equal at least 50 if our aim is to keep inbreeding rate below 1%. Still, even if inbreeding rate is 1%, the loss of genetic diversity is appreciable after a few generations, and a gradual erosion of genetic variation cannot be avoided. Eventually, the population will become virtually homozygous, the time depending on
Ne. Consequently, 1% criterion must be viewed as short-term criterion. A population with an effective size of 50 will lose about a quarter of its genetic diversity after 20–30 generations, and along with this, much of its capacity to adapt to changing conditions [
97].
To maintain a genetically healthy population in these situations,
Ne must be increased. FAO/UNEP [
95] suggests that G must approximately equal to
Ne, G being the number of generations the population is likely to retain its fitness at a relatively high level. Still, to conserve short-term fitness, or to maintain short-term fitness in captive populations other criteria must be accounted for, given effective population size is considerably affected by unbalanced sex ratios, population size evolution, by a non-random distribution of progeny among families, and other characteristics of the breeding systems implemented.
In line with these suggestions, Leroy, et al. [
98] have reported that restricting the number of generations when calculating effective size may be a good option for population monitoring, due to its effectivity to detect short-term changes in genetic diversity while it permits a generation scaled increased accuracy of the estimation of effective size, while reducing the bias related to ancestral pathways disequilibrium in pedigrees. Still, the consideration of very limited number of generations may not completely account for the lack of independence of family sizes across generations. As a result, for breeds with relatively complete pedigree records, as the ones in our study, the estimation of effective size via coancestry rate may be of interest to provide an evaluation of long-term changes in genetic diversity over long periods. For instance, the reduced numbers for effective size calculated via individual coancestry rate found in PRE, can be linked to the recent sharp bottleneck occurring in 2008 as a direct consequence of the economic crisis (
Figure 1), as afterwards, there may have been an overrepresentation of popular individuals, which drastically increased individual increase in inbreeding rate and whose effects may still be present in the current PRE population.
As suggested by Gutiérrez et al. [
99], valuable information concerning population structure can be inferred from the comparison between individual increase in inbreeding and coancestry rate pathways to compute effective population sizes. This occurs as the two parameters are assumed to be measures of the same accumulated drift process, from the founding population to the present time. For instance, as reported by Malhado, et al. [
100] both measurements of effective population size would be asymptotically equivalent in an idealized population and the disagreement between them may be mainly caused by their differential ability to assess the effect of preferential mating.
The imbalance between effective population size calculated via individual inbreeding and via coancestry rate suggests that the introduction of new reproductive individuals and the design of a plan for recommendable breeding pairs within the breeds populations could be a viable alternative to maintain the lowest possible ΔR coefficients, thereby preventing the increase in the probability of inbreeding deleterious effects from occurring and increasing the future genetic variability and effective size of the breeds.
Contextually, Alderson [
82] would suggest that, apart from controlling inbreeding to develop genetic management program in animal populations, the consideration of the GCI in breeding programs focuses and seeks to maximize the retention of the allelic richness present in the base population, but it does not take into account pedigree bottlenecks which may have occurred through non-founder individuals. In these regards, Sørensen et al. [
101] demonstrated that the comparison of
fe and
fa could be used to assess the occurrence of changes in genetic drift and recent bottlenecks in a population, which are corroborated if (
fe/fa)>1, respectively. In our study,
fe and
fa were 2.35, 1.54, and 1.48, respectively, for each of the horse breeds considered (PRá, PRE, and Há). This finding is indicative of the fact that genetic drift may not have been stable in the three horse breeds studied, with a progressive loss of founder representation, which had previously been reported in the literature [
93,
101]. Despite these values for
fe/f
a, our study confirms that an increasing trend has been described by GCI over time. Hence, the fact that founder representation is being progressively gained, not only in the Há horse breed, but also in its two ancestor breeds, may derive from the attempts of breeders and breeding associations to plan matings, trying to compensate for the aforementioned loss of founder representation.