Occurrence of Antimicrobial Resistance Genes in the Oral Cavity of Cats with Chronic Gingivostomatitis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects Included and Study Design
2.2. Sample Collection, DNA Extraction, Preparation, and Sequencing
2.3. Detection of AMR Genes, and Correlation Analysis with Malassezia restricta
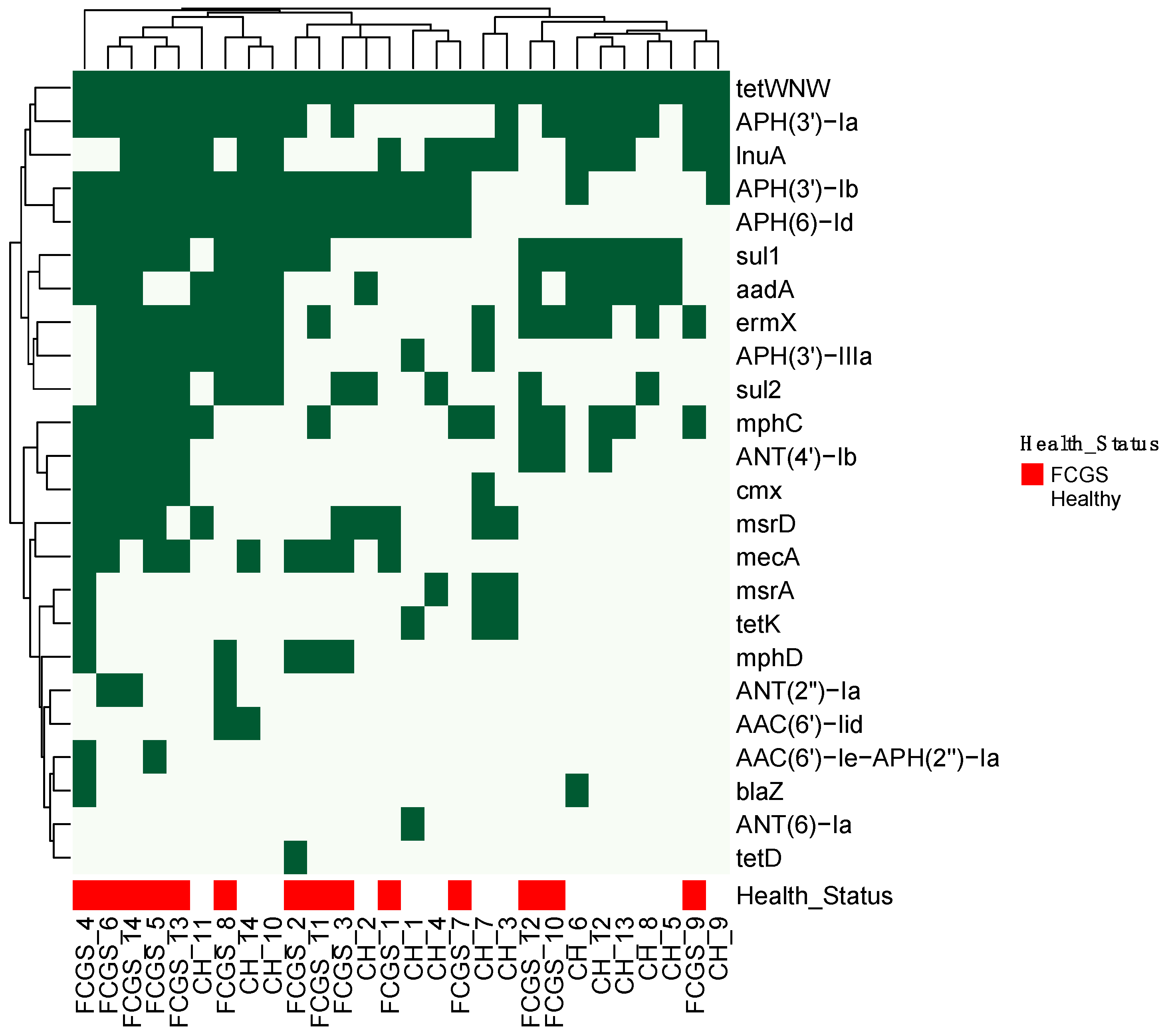
3. Results
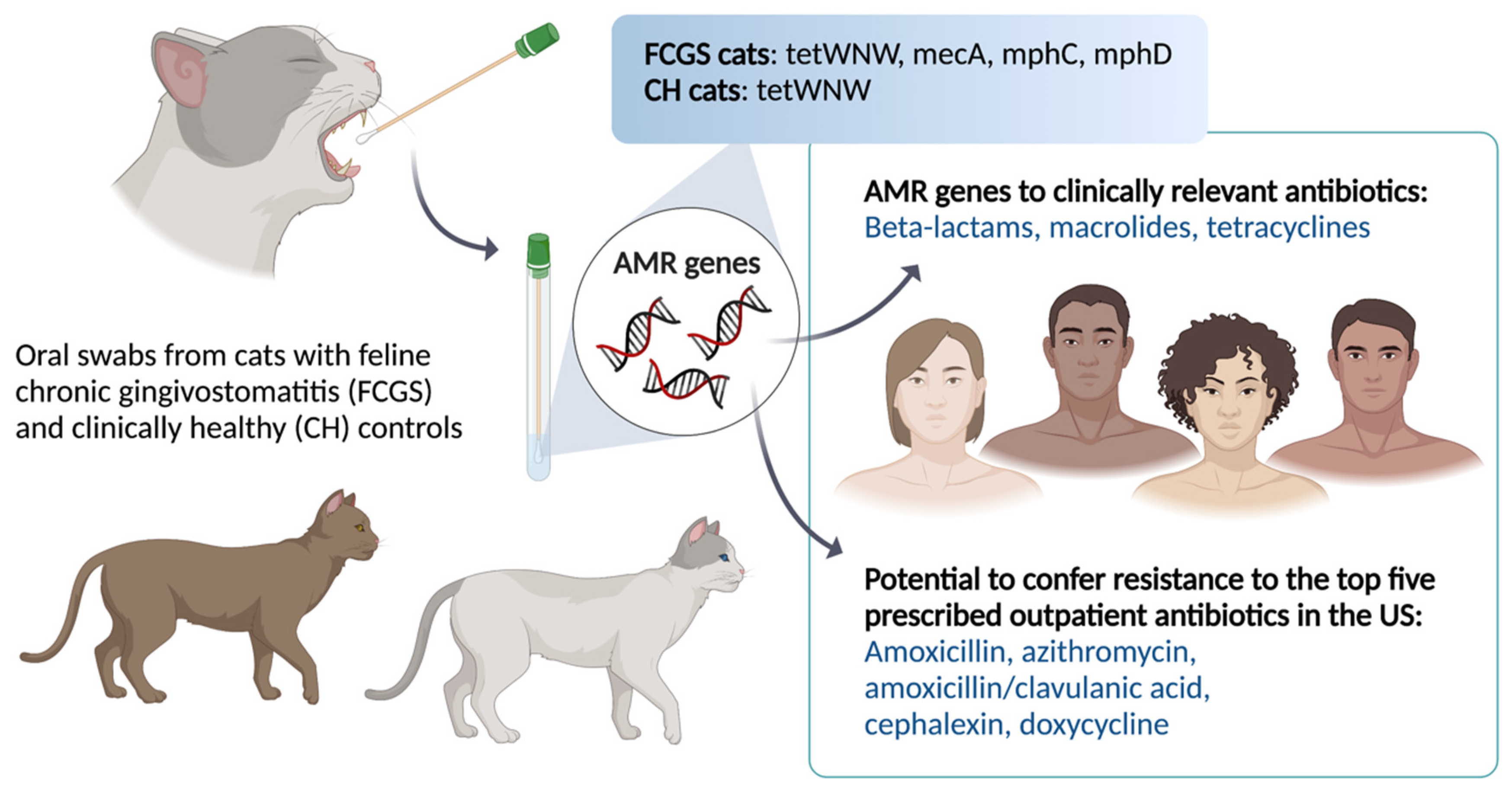
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, D.B.; Verstraete, F.J.; Arzi, B. An Update on Feline Chronic Gingivostomatitis. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 973–982. [Google Scholar] [CrossRef]
- Könönen, E.; Gursoy, M.; Gursoy, U.K. Periodontitis: A Multifaceted Disease of Tooth-Supporting Tissues. J. Clin. Med. 2019, 8, 1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winer, J.N.; Arzi, B.; Verstraete, F.J.M. Therapeutic Management of Feline Chronic Gingivostomatitis: A Systematic Review of the Literature. Front. Vet. Sci. 2016, 3, 54. [Google Scholar] [CrossRef]
- Krumbeck, J.A.; Reiter, A.; Pohl, J.; Tang, S.; Kim, Y.; Linde, A.; Prem, A.; Melgarejo, T. Characterization of Oral Microbiota in Cats: Novel Insights on the Potential Role of Fungi in Feline Chronic Gingivostomatitis. Pathogens 2021, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Spatz, M.; Richard, M.L. Overview of the Potential Role of Malassezia in Gut Health and Disease. Front. Cell. Infect. Microbiol. 2020, 10, 201. [Google Scholar] [CrossRef]
- Dupuy, A.K.; David, M.S.; Li, L.; Heider, T.N.; Peterson, J.D.; Montano, E.A.; Dongari-Bagtzoglou, A.; Diaz, P.I.; Strausbaugh, L.D. Redefining the Human Oral Mycobiome with Improved Practices in Amplicon-based Taxonomy: Discovery of Malassezia as a Prominent Commensal. PLoS ONE 2014, 9, e90899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, M.X.; Bicalho, R.C.; Fiani, N.; Lima, S.F.; Peralta, S. The subgingival microbial community of feline periodontitis and gingivostomatitis: Characterization and comparison between diseased and healthy cats. Sci. Rep. 2019, 9, 12340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meason-Smith, C.; Diesel, A.; Patterson, A.P.; Older, C.E.; Johnson, T.J.; Mansell, J.M.; Suchodolski, J.S.; Hoffmann, A.R. Characterization of the cutaneous mycobiota in healthy and allergic cats using next generation sequencing. Vet. Dermatol. 2017, 28, 84–94. [Google Scholar]
- Rolim, V.M.; Pavarini, S.P.; Campos, F.S.; Pignone, V.; Faraco, C.; de Souza Muccillo, M.; Roehe, P.M.; da Costa, F.V.A.; Driemeier, D. Clinical, pathological, immunohistochemical and molecular characterization of feline chronic gingivostomatitis. J. Feline Med. Surg. 2017, 19, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Prem, A.; Tjokrosurjo, J.; Sary, M.; van Bel, M.A.; Rodrigues-Hoffmann, A.; Kavanagh, M.; Wu, G.; van Eden, M.E.; Krumbeck, J.A. The Canine Skin and Ear Microbiome: A Comprehensive Survey of Pathogens Implicated in Canine Skin and Ear Infections Using a Novel Next-Generation-Sequencing-Based Assay. Vet. Microbiol. 2020, 247, 108764. [Google Scholar] [CrossRef]
- CARD. The Comprehensive Antibiotic Resistance Database. McMaster University: Hamilton, ON, Canada; Available online: https://card.mcmaster.ca/ontology/41634 (accessed on 25 October 2021).
- Lyon, K.F. Gingivostomatitis. Vet. Clin. N. Am. Small Anim. Pract. 2005, 35, 891–911. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, B.E.; Griffin, C.E.; Rosenkrantz, W.S. Response of feline eosinophilic plaques and lip ulcers to amoxicillin trihydrate-clavulanate potassium therapy: A randomized, double-blind placebo-controlled prospective study. Vet. Dermatol. 2011, 23, e24–e25. [Google Scholar] [CrossRef] [PubMed]
- Durkin, M.J.; Jafarzadeh, S.R.; Hsueh, K.; Sallah, Y.H.; Munshi, K.D.; Henderson, R.R.; Fraser, V.J. Outpatient Antibiotics Prescription Trends in the United States: A National Cohort Study. Infect. Control Hosp. Epidemiol. 2018, 39, 584–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–217. [Google Scholar] [CrossRef]
- Biswas, S.; Maggi, R.; Papich, M.; Keil, D.; Breitschwerdt, E. Comparative Activity of Pradofloxacin, Enrofloxacin, and Azithromycin against Bartonella henselae Isolates Collected from Cats and a Human. J. Clin. Microbiol. 2009, 48, 2. [Google Scholar] [CrossRef] [Green Version]
- Ruch-Gallie, R.; Veir, K.; Spindel, M.; Lappin, M. Efficacy of amoxycillin and azithromycin for the empirical treatment of shelter cats with suspected bacterial upper respiratory infections. J. Feline Med. Surg. 2008, 10, 542–550. [Google Scholar] [CrossRef]
- Cohn, L.A.; Birkenheuer, A.J.; Brunker, J.D.; Ratcliff, E.R.; Craig, A.W. Efficacy of Atovaquone and Azithromycin or Imidocarb Dipropionate in Cats with Acute Cytauxzoonosis. J. Vet. Intern. Med. 2011, 25, 25–60. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Outpatient Antibiotics Prescriptions. Available online: https://www.cdc.gov/antibiotic-use/pdfs/Annual-Report-2020-H.pdf (accessed on 25 October 2021).
- The World Health Organization. Critically Important Antimicrobials for Human Medicine. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 25 October 2021).
- Plumb, D.C. Aminoglycosides. In Plumb’s Veterinary Drug Handbook: Desk, 9th ed.; P Wiley-Blackwell: Hoboken, NJ, USA, 2018; pp. 346–364. [Google Scholar]
- Del Prete, S.; Vullo, D.; Ghobril, C.; Hitce, J.; Clavaud, C.; Marat, X.; Clemente, C.; Supuran, C.T. Cloning, purification, and characterization of a β-carbonic anhydrase from Malassezia restricta, an opportunistic pathogen involved in dandruff and seborrheic dermatitis. Int. J. Mol. Sci. 2019, 20, 2447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stalhberger, T.; Simenel, C.; Clavaud, C.; Eijsink, V.G.; Jourdain, R.; Delepierre, M.; Latgé, J.P.; Breton, L.; Fontaine, T. Chemical organization of the cell wall polysaccharide core of Malassezia restricta. J. Biol. Chem. 2014, 289, 12647–12656. [Google Scholar] [CrossRef] [Green Version]
- Morand, S.C.; Bertignac, M.; Iltis, A.; Kolder, I.C.R.M.; Pirovano, W.; Jourdain, R.; Clavaud, C. Complete Genome Sequence of Malassezia restricta CBS 7877, an Opportunist Pathogen Involved in Dandruff and Seborrheic Dermatitis. Microbiol. Resour. Announc. 2019, 8, e01543-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamilselvi, A.; Mugesh, G. Zinc and antibiotic resistance: Metallo-beta-lactamases and their synthetic analogues. J. Biol. Inorg. Chem. 2008, 13, 1039–1053. [Google Scholar] [CrossRef] [PubMed]
| AMR Gene | Antibiotic Resistance | CH | FCGS | z-Score | p-Value |
|---|---|---|---|---|---|
| aph33Ib | Aminoglycosides | 8 | 11 | −1.22 | 0.22 |
| aph3Ia | Aminoglycosides | 9 | 10 | −0.40 | 0.69 |
| aph6Id | Aminoglycosides | 6 | 11 | −1.93 | 0.05 |
| ant2Ia | Aminoglycosides | 0 | 3 | −1.83 | 0.07 |
| ant4Ib | Aminoglycosides | 1 | 7 | −2.51 | 0.01 * |
| aac6IId | Aminoglycosides | 1 | 1 | 0.00 | 1.00 |
| aac6Ie | Aminoglycosides | 0 | 2 | −1.47 | 0.14 |
| aadA | Aminoglycosides | 9 | 5 | 1.51 | 0.13 |
| ant6Ia | Aminoglycosides | 1 | 0 | 1.02 | 0.31 |
| aph3IIIa | Aminoglycosides | 5 | 5 | 0.00 | 1.00 |
| mecA | Beta-Lactams | 1 | 8 | −2.83 | 0.00 * |
| blaZ | Beta-Lactams | 1 | 1 | 0.00 | 1.00 |
| cmx | Florfenicols | 1 | 5 | −1.85 | 0.06 |
| sul1 | Sulfonamides | 7 | 10 | −1.16 | 0.25 |
| sul2 | Sulfonamides | 6 | 7 | −0.76 | 0.45 |
| mphD | Macrolides | 0 | 5 | −2.47 | 0.01 * |
| mphC | Macrolides | 4 | 10 | 2.26 | 0.02 * |
| lnuA | Lincosamides | 10 | 6 | 1.52 | 0.13 |
| tetD | Tetracyclines | 0 | 1 | −1.02 | 0.31 |
| tetWNW | Tetracyclines | 14 | 14 | 0.00 | 1.00 |
| tetK | Tetracyclines | 3 | 1 | 1.08 | 0.28 |
| ermX | Macrolides, Lincosamides | 7 | 9 | −0.76 | 0.45 |
| msrA | Macrolides, Tetracyclines, Lincosamides | 3 | 1 | 1.08 | 0.28 |
| msrD | Macrolides, Tetracyclines, Lincosamides | 4 | 6 | −0.79 | 0.43 |
| AMR Gene | Correlation Coefficient | p-Value |
|---|---|---|
| mecA | 0.49381 | 0.00757 * |
| mphD | 0.62317 | 0.0004 * |
| mphC | −0.07606 | 0.70048 |
| Ant4lb | 0.08136 | 0.68066 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsang, W.; Linde, A.; Krumbeck, J.A.; Wu, G.; Kim, Y.J.; Lushington, G.H.; Melgarejo, T. Occurrence of Antimicrobial Resistance Genes in the Oral Cavity of Cats with Chronic Gingivostomatitis. Animals 2021, 11, 3589. https://doi.org/10.3390/ani11123589
Tsang W, Linde A, Krumbeck JA, Wu G, Kim YJ, Lushington GH, Melgarejo T. Occurrence of Antimicrobial Resistance Genes in the Oral Cavity of Cats with Chronic Gingivostomatitis. Animals. 2021; 11(12):3589. https://doi.org/10.3390/ani11123589
Chicago/Turabian StyleTsang, Wayne, Annika Linde, Janina A. Krumbeck, Guangxi Wu, Young J. Kim, Gerald H. Lushington, and Tonatiuh Melgarejo. 2021. "Occurrence of Antimicrobial Resistance Genes in the Oral Cavity of Cats with Chronic Gingivostomatitis" Animals 11, no. 12: 3589. https://doi.org/10.3390/ani11123589
APA StyleTsang, W., Linde, A., Krumbeck, J. A., Wu, G., Kim, Y. J., Lushington, G. H., & Melgarejo, T. (2021). Occurrence of Antimicrobial Resistance Genes in the Oral Cavity of Cats with Chronic Gingivostomatitis. Animals, 11(12), 3589. https://doi.org/10.3390/ani11123589






