Host Identity and Geographic Location Significantly Affect Gastrointestinal Microbial Richness and Diversity in Western Lowland Gorillas (Gorilla gorilla gorilla) under Human Care
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. Amplicon Sequencing
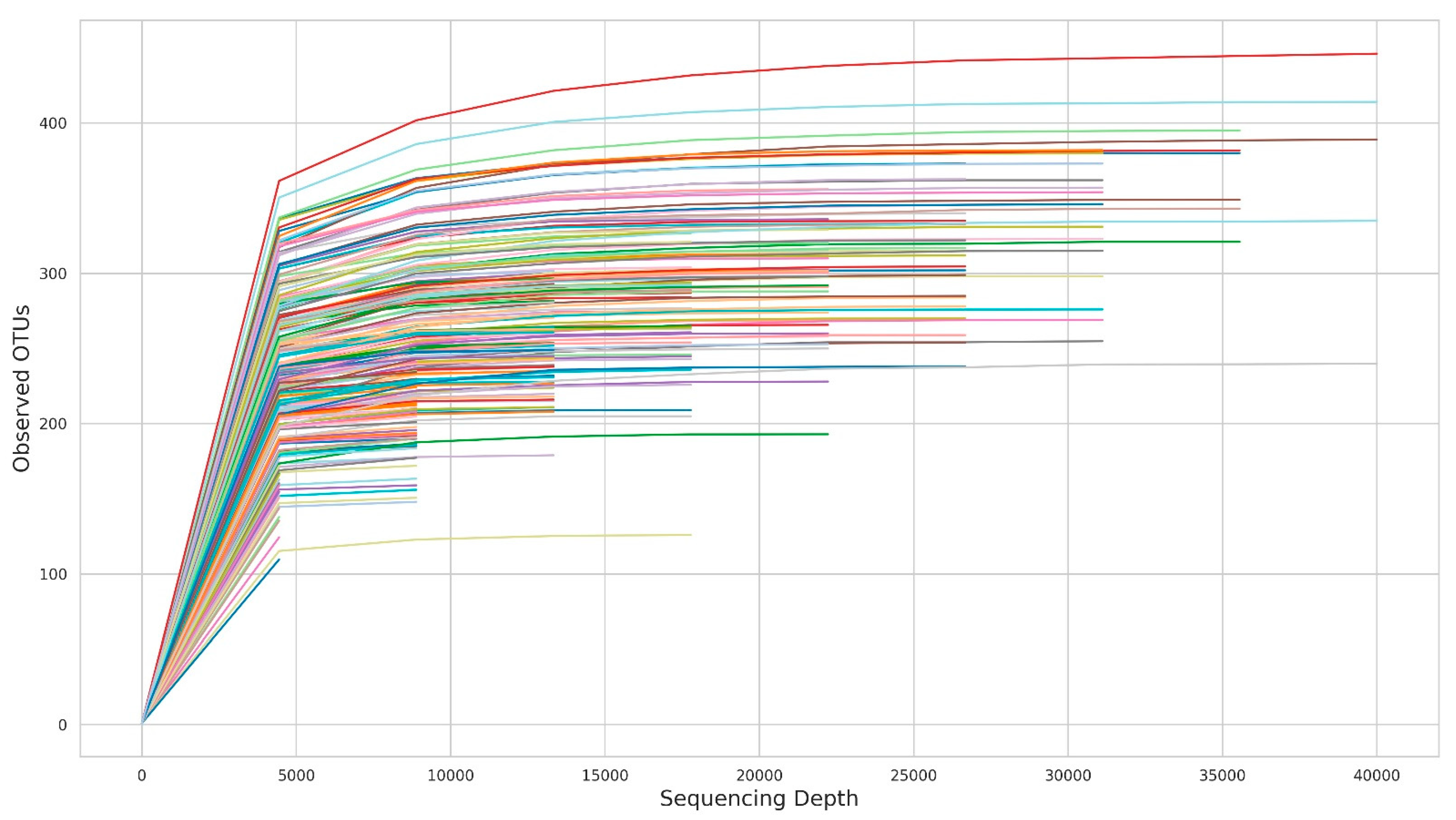
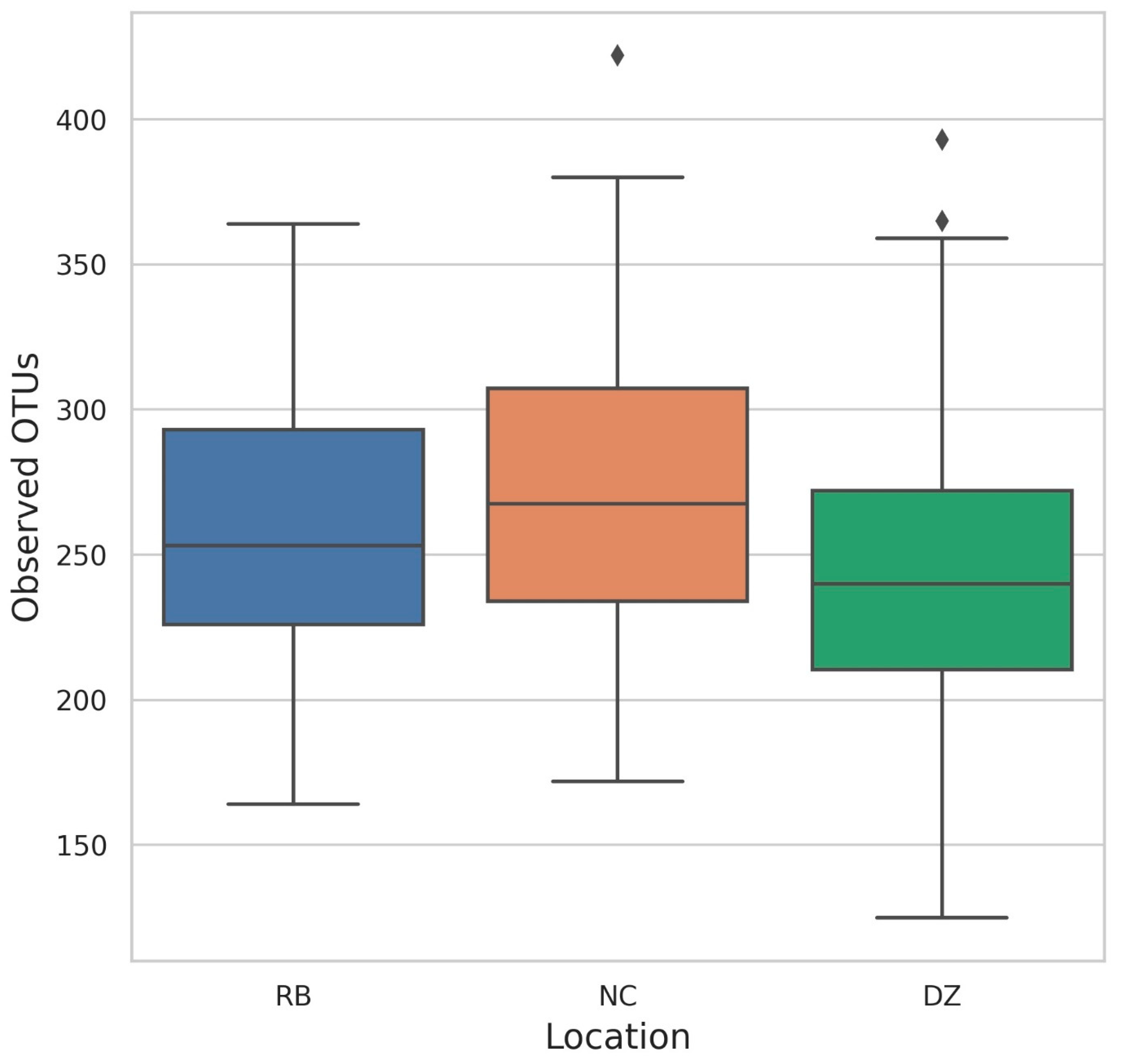
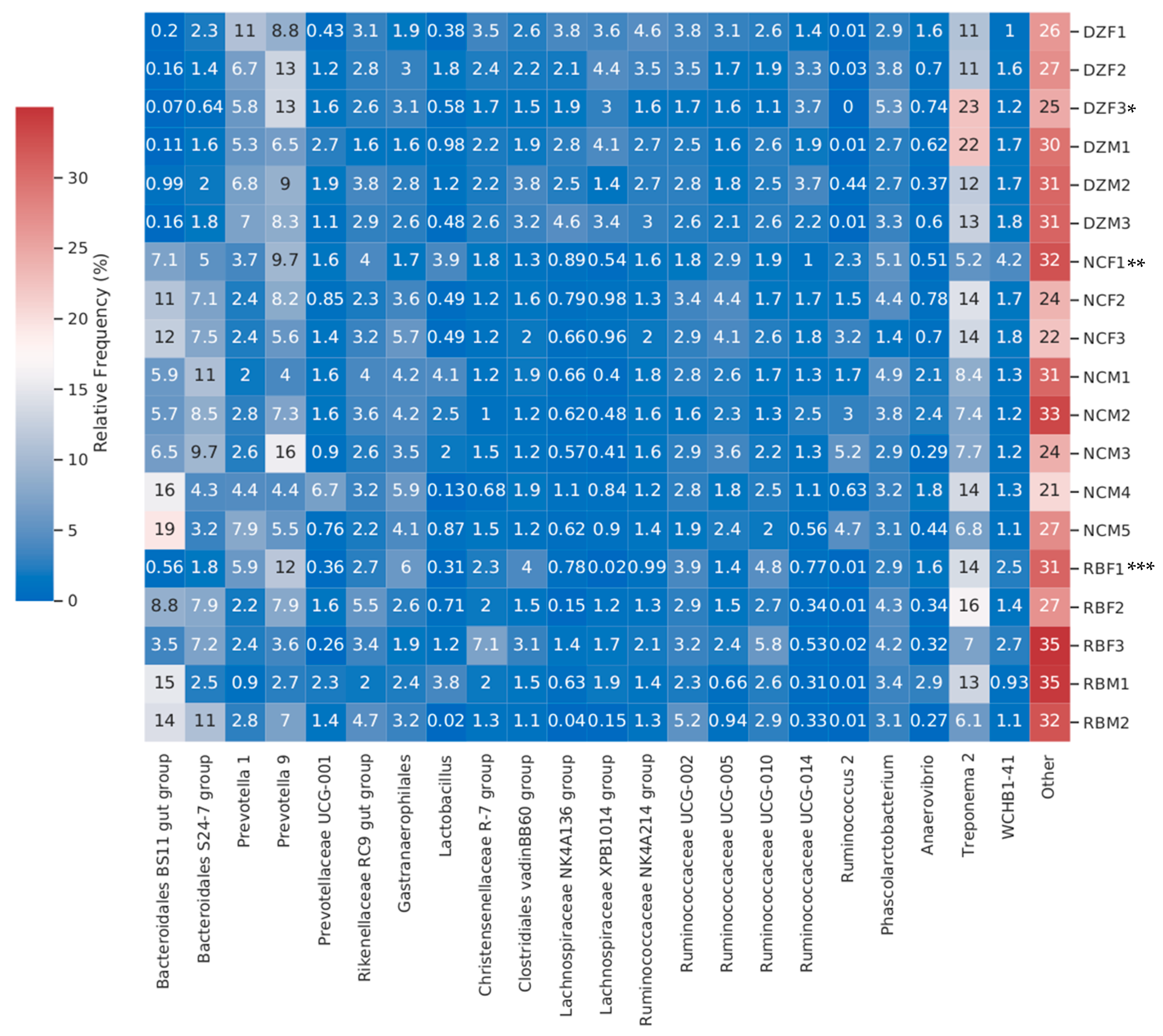
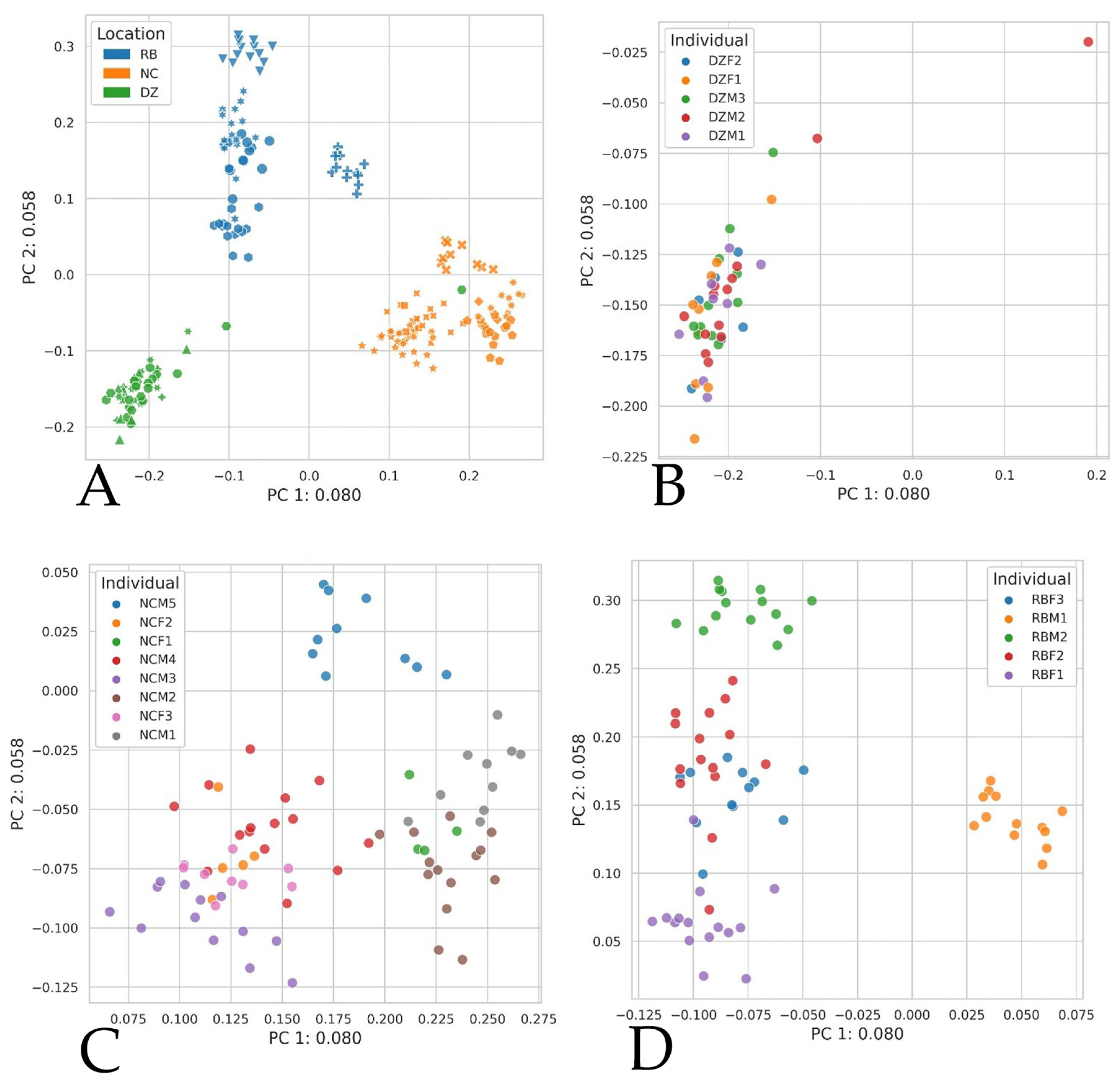
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Amato, K.R.; Martinez-Mota, R.; Righini, N.; Raguet-Schofield, M.; Corcoine, F.P.; Marini, E.; Humphrey, G.; Gogul, G.; Gaffney, J.; Lovelace, E.; et al. Phylogenetic and Ecological Factors Impact the GI Microbiota of two Neotropical Primate Species. Oecologia 2016, 180, 717–733. [Google Scholar] [CrossRef]
- Amato, K.; Mallot, K.; McDonald, D.; Dominy, N.; Goldberg, T.; Lambert, J.; Swedell, L.; Metcalf, J.; Gomez, A.; Britton, G.; et al. Convergence of Human and Old World Monkey Gut Microbiomes Demonstrates the Importance of Human Ecology over Phylogeny. Genome Biol. 2019, 20, 201. [Google Scholar] [CrossRef] [PubMed]
- Clayton, J.B.; Vangay, P.; Huang, H.; Ward, T.; Hillmann, B.M.; Al-Ghalith, G.A.; Travis, D.A.; Long, H.T.; Tuan, B.V.; Minh, V.V.; et al. Captivity Humanizes the Primate Microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 10376–10381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, L.K.; McKenney, E.A.; O’Connell, T.M.; Drea, C.M. The Critical Role of Dietary Foliage in Maintaining the GI Microbiome and Metabolome of Folivorous Sifakas. Nature 2018, 8, 14482. [Google Scholar]
- Greene, L.K.; Clayton, J.B.; Rothman, R.S.; Semel, B.P.; Semel, M.A.; Gillespie, T.R.; Wright, P.C.; Drea, C.M. Local Habit, not Phylogenetic Relatedness, Predicts Gut Microbiota Better within Folivorous than Frugivorous Lemur Lineages. Biol. Lett. 2019, 15, 20190028. [Google Scholar] [CrossRef] [PubMed]
- Griffen, J.L.; Wang, X.; Stanley, E. Does our GI Microbiome Predict Cardiovascular Risk? A Review of the Evidence from Metabolomics. Circ. Cardiovasc. Genet. 2015, 8, 187–191. [Google Scholar] [CrossRef] [Green Version]
- McKenney, E.A.; Ashwell, M.; Lambert, J.E.; Fellner, V. Fecal Microbial Diversity and Putative Function in Captive Western Lowland Gorillas (Gorilla gorilla gorilla), Common Chimpanzees (Pan troglodytes), Hamadryas Baboons (Papio hamadryas) and Binturongs (Arctictis binturong). Integr. Zool. 2014, 9, 557–569. [Google Scholar] [CrossRef] [PubMed]
- McKenney, E.A.; Rodrigo, A.; Yoder, A.D. Patterns of GI Bacterial Colonization in Three Primate Species. PLoS ONE 2015, 10, e0124618. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, V.J.; Song, S.J.; Delsuc, F.; Prest, T.L.; Oliverio, A.M.; Korpita, T.M.; Alexiev, A.; Amato, K.R.; Metcalf, J.L.; Kowalewski, M.; et al. The Effects of Captivity on the Mammalian GI Microbiome. Integr. Comp. Biol. 2017, 4, 690–704. [Google Scholar] [CrossRef] [Green Version]
- Gomez, A.; Rothman, J.M.; Petrzelkova, K.; Yeoman, C.J.; Vlckova, K.; Umaña, J.D.; Carr, M.; Modry, D.; Todd, A.; Torralba, M.; et al. Temporal variation selects for diet–microbe co-metabolic traits in the gut of Gorilla spp. ISME J. 2016, 10, 514–526. [Google Scholar] [CrossRef] [Green Version]
- Campbell, T.P.; Sun, X.; Patel, V.H.; Sanz, C.; Morgan, D.; Dantas, G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020, 14, 1584–1599. [Google Scholar] [CrossRef] [PubMed]
- Narat, V.; Amato, K.R.; Ranger, N.; Salmona, M.; Mercier-Delarue, S.; Rupp, S.; Ambata, P.; Njouom, R.; Simon, F.; Giles-Vernick, T.; et al. A multi-disciplinary comparison of great ape gut microbiota in a central African forest and European zoo. Scient. Rep. 2020, 10, 19107. [Google Scholar] [CrossRef]
- Springer, A.; Fichtel, C.; Al-Ghalith, G.A.; Koch, F.; Amato, K.R.; Clayton, J.B.; Knights, D.; Kappeler, P.M. Patterns of Seasonality and Group Membership Characterize the GI Microbiota in a Longitudinal Study of Wild Verreaux’s Sifakas (Propithecus verreauxi). Ecol. Evol. 2017, 7, 15732–15745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochman, H.; Worobey, M.; Kuo, C.; Ndjango, J.N.; Peeters, M.; Hahn, B.H.; Hugenholtz, P. Evolutionary Relationships of Woild Hominids Recapitulated by GI Microbial Communities. PLoS Biol. 2010, 8, e1000546. [Google Scholar] [CrossRef]
- Yildirim, S.; Yeoman, C.J.; Sipos, M.; Torralba, M.; Wilson, B.A.; Goldberg, T.L.; Stumpf, R.M.; Leigh, S.R.; White, B.A.; Nelson, K.E. Characterization of the Fecal Microbiome from Non-Human Primates Reveals Species Specific Microbial Communities. PLoS ONE 2010, 5, e13963. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.T.; Peek, R.M., Jr. GI Malignancy and the Microbiome. Gastroenterology 2014, 146, 1534–1546. [Google Scholar] [CrossRef] [Green Version]
- Hollister, E.B.; Gao, C.; Versalovic, J. Compositional and Functional Features of the GI Microbiome and Their Effects on Human Health. Gastroenterology 2014, 146, 1449–1458. [Google Scholar] [CrossRef] [Green Version]
- McKenney, E.A.; Greene, L.K.; Drea, C.M.; Yoder, A.D. Down for the Count: Cryptosporidium Infection Depletes the GI Microbiome in Coquerel’s Sifakas. Microb. Ecol. Health Dis. 2017, 28, 1335165. [Google Scholar] [CrossRef] [Green Version]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The Long-Term Stability of the Human GI Microbiota. Science 2013, 340, 44–48. [Google Scholar]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Jimenez, B.; Wilkinson, M.D. Robust and Automatic Definition of Microbiome States. PeerJ 2019, 7, e6657. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.K.; Costella, E.K.; Nemergut, D.R.; Cleveland, C.C.; Reed, S.C.; Weintraub, M.N.; Meyer, A.F.; Martin, A.M. Biogeochemical Consequences of Rapid Microbial Turnover and Seasonal Succession in Soil. Ecoloby 2007, 88, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Bornbusch, S.L.; Greene, L.K.; Rahobilalaina, S.; Calkins, S.; Clarke, T.A.; LaFleur, M.; Drea, C.M. Gut Microbiota of Ring-Tailed Lemurs (Lemur catta) Vary Across Natural and Captive Populations and Correlate with Environmental Microbiota. bioRxiv 2021, 1–49. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [Green Version]
- Gotelli, N.J.; Colwell, R.K. Estimating Species Richness. In Biological Diversity: Frontiers in Measurement and Assessment; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vich Vila, A.; Falony, G.; Vieira-Silva, S.; et al. Population-Based Metagenomics Analysis Reveals Markers for Gut Microbiome Composition and Diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-4, 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 March 2021).
- Knights, D. Discovering Patterns in the Microbiome. University of Minnesota. 2016. Available online: Metagenome.cs.umn.edu/microbiomecodebrowser/doc/index.html (accessed on 10 August 2021).
- Montassier, E.; Al-Ghalith, G.A.; Hillmann, B.; Visocil, K.; Kabage, A.J.; McKinlay, C.E.; Sadowsky, M.J.; Khoruts, A.; Knights, D. CLOUD: A Non-Parametric Detection Test for Microbiome Outliers. Microbiome 2018, 6, 137. [Google Scholar] [CrossRef]
- Randolf, J.D. Treponema. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galeston: Galveston, TX, USA, 1996; Available online: https://www.ncbi.nlm.nih.gov/books/NBK7716/ (accessed on 25 September 2021).
- Tung, J.; Barreiro, L.B.; Burns, M.B.; Grenier, J.C.; Lynch, J.; Grieneisen, L.E.; Altmann, J.; Alberts, S.C.; Blekhman, R.; Archie, E.A. Social Networks Predict GI Microbiome Composition in Wild Baboons. eLife 2015, 4, e05224. [Google Scholar] [CrossRef]
- Zhu, L.; Clayton, J.B.; Suhr Van Haute, M.J.; Yang, Q.; Hassenstab, H.R.; Mustoe, A.C.; Knights, D.; Benson, A.K.; French, J.A. Sex Bias in Gut Microbiome Transmission in Newly Paired Marmosets (Callithrix jacchus). Am. Soc. Microbiol. 2020, 5, e00910-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, A.; Petrzelkova, K.; Yeoman, C.J.; Vlkova, K.; Mrazek, J.; Koppova, I.; Carbonero, F.; Ulanov, A.; Modry, D.; Todd, A.; et al. GI Microbiome Composition and Metabolomics Profiles of Wild Western Lowland Gorillas (Gorilla gorilla gorilla) Reflect Host Ecology. Mol. Ecol. 2015, 24, 2551–2565. [Google Scholar] [CrossRef]
- Clayton, J.B.; Al-Ghalith, G.A.; Long, H.T.; Tuan, B.V.; Cabana, F.; Huang, H.; Vangay, P.; Ward, T.; Minh, V.V.; Tam, N.A.; et al. Associations Between Nutrition, GI Microbiome, and Health in a Novel Non-Human Primate Model. Scient. Rep. 2018, 8, 11159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krynak, K.L.; Burke, D.J.; Martin, R.A.; Dennis, P.M. Gut Microbiome Composition is Associated with Cardiac Disease in Zoo-Housed Western Lowland Gorillas (Gorilla gorilla gorilla). FEMS Microbiol. Lett. 2017, 364, 15. [Google Scholar] [CrossRef]
- Hao, L.; Meier-Kolthoff, J.P.; Hu, C.; Wang, Z.; Zhu, J.; Zheng, W.; Tian, Y.; Guo, F. Panoramic Insights into the Microevolution and Macroevolution of Prevotella copri- Containing Lineage in Primate Guts. bioRxiv 2020, 1–55. [Google Scholar] [CrossRef]
- Moeller, A.H.; Peeters, M.; Ndjango, J.B.; Li, Y.; Hahn, B.H.; Ochman, H. Sympatric Chimpanzees and Gorillas Harbor Convergent GI Microbial Communities. Genome 2013, 23, 1715–1720. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanuja, S.P.S.; Shasany, A.K.; Kumar, S. Rapid Isolation of DNA from Dry and Fresh Samples of Plants Producing Large Amounts of Secondary Metabolites and Essential Oils. Plant Mol. Biol. Report. 1999, 17, 74. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]

| Institution 1 | Leafy Green Vegetables | Celery 2 | Other Vegetables | Fruit | Primate Biscuit 3 | Browse | Alfalfa |
|---|---|---|---|---|---|---|---|
| DZ | 40 | 11.7 | 20 | na | 1.8 | 15 | 11.5 |
| NC | 66 | na | 27 | 7.0 | na | variable | na |
| RB | 60 | na | 33 | 7.0 | na | variable | na |
| Animal ID 1–3 | Total Samples Collected | Number of Samples after Rarefaction | Housing Group 4 |
|---|---|---|---|
| DZM1 | 16 | 8 | DZ1 |
| DZM2 | 17 | 13 | DZ2 |
| DZM3 | 16 | 11 | DZ2 |
| DZF1 | 16 | 9 | DZ1 |
| DZF2 | 16 | 6 | DZ1 |
| DZF3 * | 3 | 0 | DZ1 |
| NCM1 | 12 | 10 | NC1 |
| NCM2 | 15 | 14 | NC1 |
| NCM3 | 13 | 13 | NC1 |
| NCM4 | 15 | 15 | NC1 |
| NCM5 | 11 | 10 | NC1 |
| NCF1 | 7 | 4 | NC1 |
| NCF2 | 8 | 5 | NC1 |
| NCF3 | 11 | 9 | NC1 |
| RBM1 | 14 | 13 | RB1 |
| RBM2 | 15 | 14 | RB2 |
| RBF1 | 15 | 15 | RB2 |
| RBF2 | 15 | 15 | RB2 |
| RBF3 | 13 | 12 | RB2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eschweiler, K.; Clayton, J.B.; Moresco, A.; McKenney, E.A.; Minter, L.J.; Suhr Van Haute, M.J.; Gasper, W.; Hayer, S.S.; Zhu, L.; Cooper, K.; et al. Host Identity and Geographic Location Significantly Affect Gastrointestinal Microbial Richness and Diversity in Western Lowland Gorillas (Gorilla gorilla gorilla) under Human Care. Animals 2021, 11, 3399. https://doi.org/10.3390/ani11123399
Eschweiler K, Clayton JB, Moresco A, McKenney EA, Minter LJ, Suhr Van Haute MJ, Gasper W, Hayer SS, Zhu L, Cooper K, et al. Host Identity and Geographic Location Significantly Affect Gastrointestinal Microbial Richness and Diversity in Western Lowland Gorillas (Gorilla gorilla gorilla) under Human Care. Animals. 2021; 11(12):3399. https://doi.org/10.3390/ani11123399
Chicago/Turabian StyleEschweiler, Katrina, Jonathan B. Clayton, Anneke Moresco, Erin A. McKenney, Larry J. Minter, Mallory J. Suhr Van Haute, William Gasper, Shivdeep Singh Hayer, Lifeng Zhu, Kathryn Cooper, and et al. 2021. "Host Identity and Geographic Location Significantly Affect Gastrointestinal Microbial Richness and Diversity in Western Lowland Gorillas (Gorilla gorilla gorilla) under Human Care" Animals 11, no. 12: 3399. https://doi.org/10.3390/ani11123399
APA StyleEschweiler, K., Clayton, J. B., Moresco, A., McKenney, E. A., Minter, L. J., Suhr Van Haute, M. J., Gasper, W., Hayer, S. S., Zhu, L., Cooper, K., & Ange-van Heugten, K. (2021). Host Identity and Geographic Location Significantly Affect Gastrointestinal Microbial Richness and Diversity in Western Lowland Gorillas (Gorilla gorilla gorilla) under Human Care. Animals, 11(12), 3399. https://doi.org/10.3390/ani11123399










