NEFA Promotes Autophagosome Formation through Modulating PERK Signaling Pathway in Bovine Hepatocytes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation, Culture, and Treatment of Calf Primary Hepatocytes
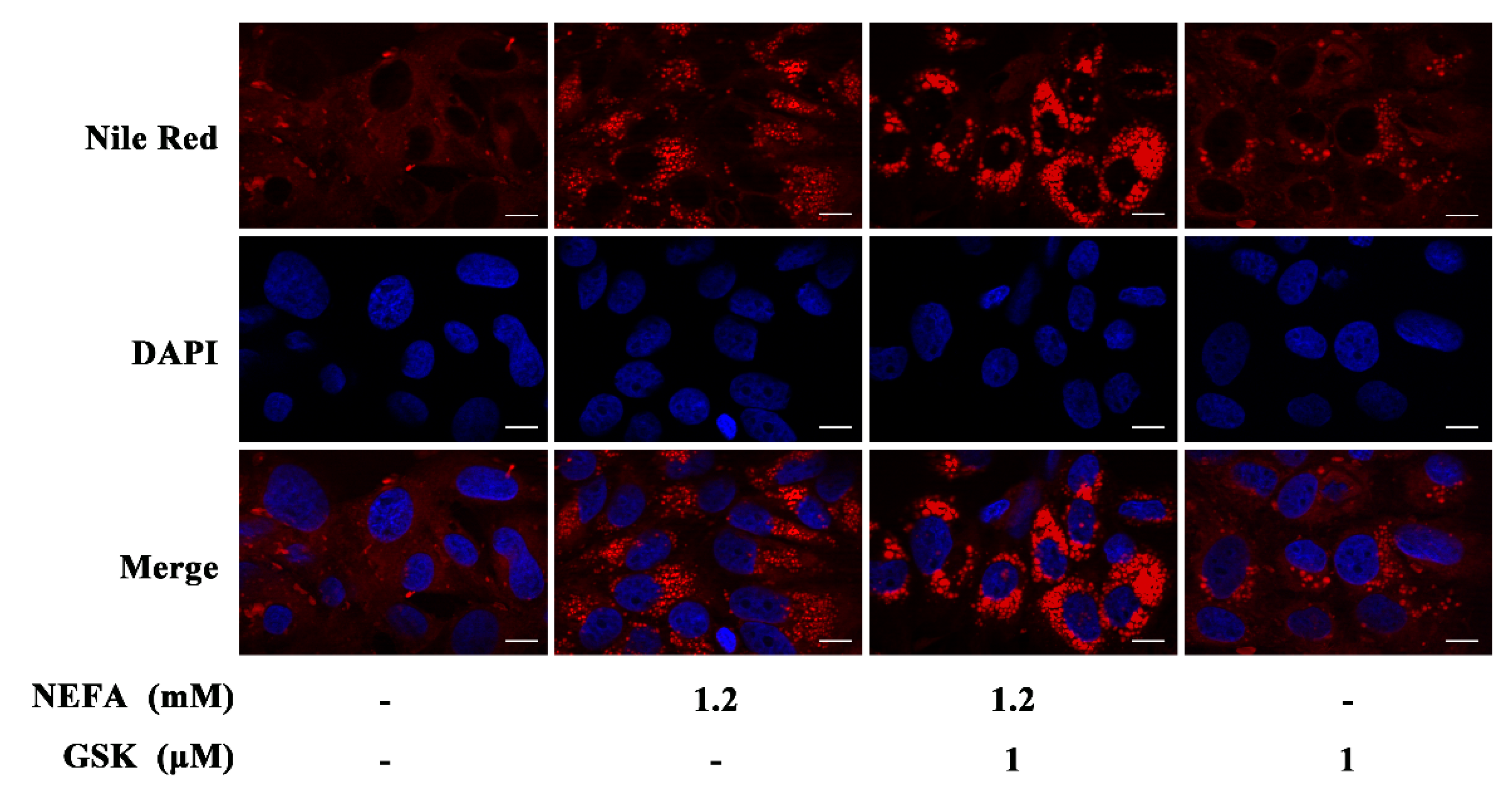
2.2. Nile Red Staining
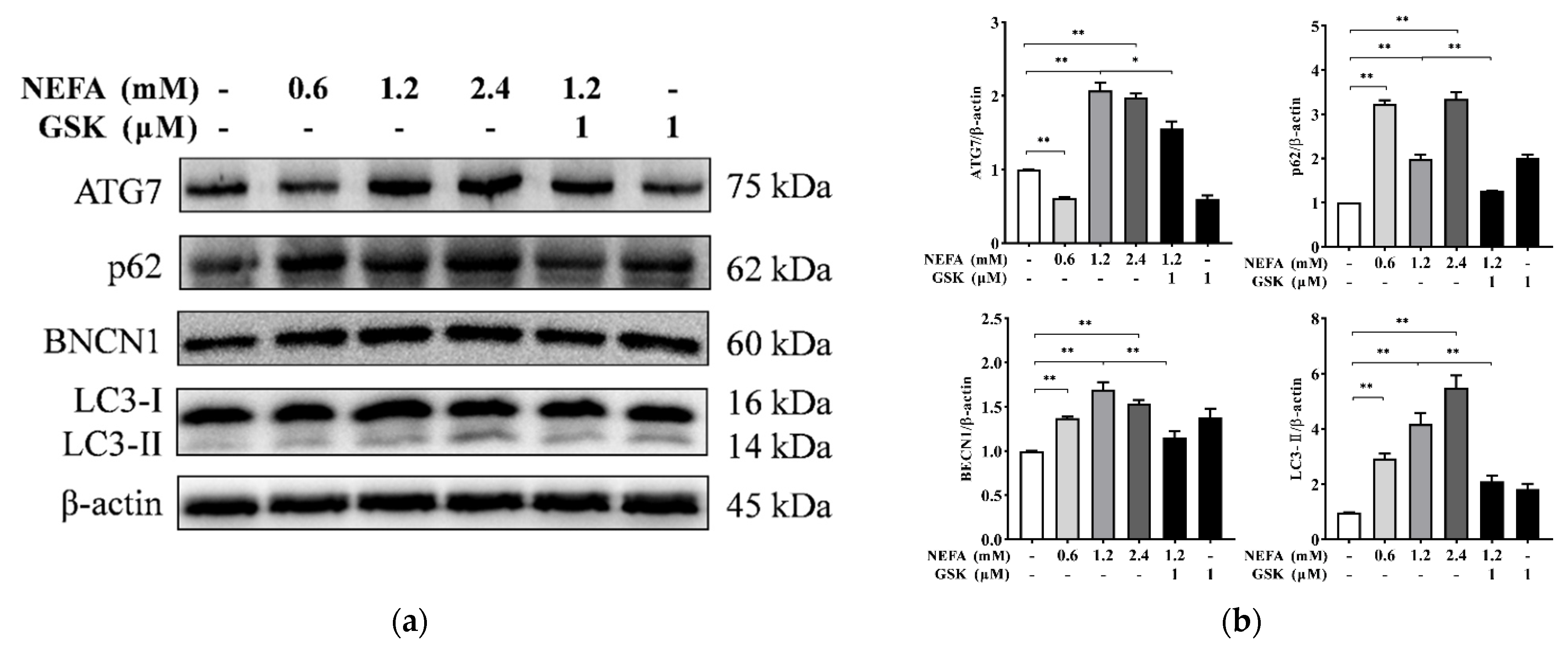
2.3. Protein Extraction and Western Blotting
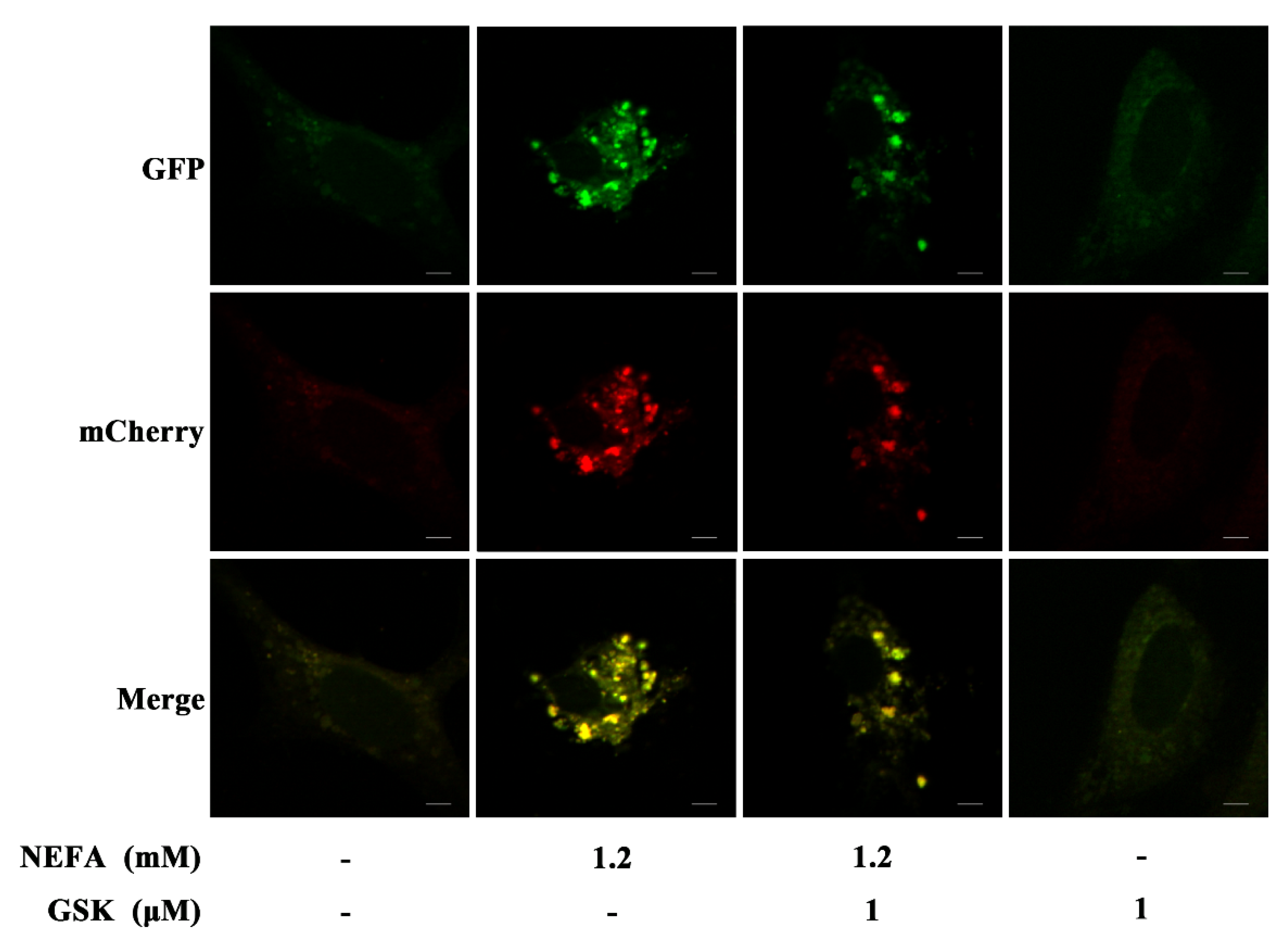
2.4. MCherry GFP LC3B Transfection
2.5. Transmission Electron Microscopy
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bell, A.W.; Bauman, D.E. Adaptations of glucose metabolism during pregnancy and lactation. J. Mammary Gland. Biol. Neoplasia. 1997, 2, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Adewuyi, A.A.; Gruys, E.; van Eerdenburg, F.J. Non esterified fatty acids (NEFA) in dairy cattle. A review. Vet. Q. 2005, 27, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.L.; Chen, S.Y.; Hu, S.; Jia, X.; Wang, J.; Lai, S.J. Metabolomic and Proteomic Profiles Associated With Ketosis in Dairy Cows. Front. Genet. 2020, 11, 551587. [Google Scholar] [CrossRef]
- Ringseis, R.; Gessner, D.K.; Eder, K. Molecular insights into the mechanisms of liver-associated diseases in early-lactating dairy cows: Hypothetical role of endoplasmic reticulum stress. J. Anim. Physiol. Anim. Nutr. 2015, 99, 626–645. [Google Scholar] [CrossRef] [PubMed]
- Dowman, J.K.; Tomlinson, J.W.; Newsome, P.N. Pathogenesis of non-alcoholic fatty liver disease. QJM Int. J. Med. 2010, 103, 71–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Zhao, C.; Kong, Y.; Tan, P.; Liu, S.; Liu, Y.; Zeng, F.; Yuan, Y.; Zhao, B.; Wang, J. Elucidation of the mechanism of NEFA-induced PERK-eIF2alpha signaling pathway regulation of lipid metabolism in bovine hepatocytes. J. Steroid Biochem. Mol. Biol. 2021, 211, 105893. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Guan, Y.; Loor, J.J.; Sha, X.; Coleman, D.N.; Zhang, C.; Du, X.; Shi, Z.; Li, X.; Wang, Z.; et al. Fatty acid-induced endoplasmic reticulum stress promoted lipid accumulation in calf hepatocytes, and endoplasmic reticulum stress existed in the liver of severe fatty liver cows. J. Dairy Sci. 2019, 102, 7359–7370. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Popko, B.; Roos, R.P. An enhanced integrated stress response ameliorates mutant SOD1-induced ALS. Hum. Mol. Genet. 2014, 23, 2629–2638. [Google Scholar] [CrossRef]
- Garcia-Esparcia, P.; Hernandez-Ortega, K.; Koneti, A.; Gil, L.; Delgado-Morales, R.; Castano, E.; Carmona, M.; Ferrer, I. Altered machinery of protein synthesis is region- and stage-dependent and is associated with alpha-synuclein oligomers in Parkinson’s disease. Acta. Neuropathol. Commun. 2015, 3, 76. [Google Scholar] [CrossRef] [Green Version]
- Meusser, B.; Hirsch, C.; Jarosch, E.; Sommer, T. ERAD: The long road to destruction. Nat. Cell Biol. 2005, 7, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Rzymski, T.; Milani, M.; Pike, L.; Buffa, F.; Mellor, H.R.; Winchester, L.; Pires, I.; Hammond, E.; Ragoussis, I.; Harris, A.L. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010, 29, 4424–4435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef]
- Chung, K.W.; Kim, K.M.; Choi, Y.J.; An, H.J.; Lee, B.; Kim, D.H.; Lee, E.K.; Im, E.; Lee, J.; Im, D.S.; et al. The critical role played by endotoxin-induced liver autophagy in the maintenance of lipid metabolism during sepsis. Autophagy 2017, 13, 1113–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Loor, J.J.; Zhai, Q.; Liang, Y.; Yu, H.; Du, X.; Shen, T.; Fang, Z.; Shi, Z.; Wang, X.; et al. Short communication: Enhanced autophagy activity in liver tissue of dairy cows with mild fatty liver. J. Dairy Sci. 2020, 103, 3628–3635. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, X.; She, G.; Kong, Y.; Guo, Y.; Wang, Z.; Liu, G.; Zhao, B. Serum hepatokines in dairy cows: Periparturient variation and changes in energy-related metabolic disorders. BMC Vet. Res. 2018, 14, 236. [Google Scholar] [CrossRef]
- Kong, Y.; Zhao, C.; Huang, Y.; Liu, Y.; Liu, S.; Guo, Y.; Li, M.; Xu, T.; Zhao, B.; Wang, J. Angiopoietin-like protein 4 promotes very-low-density lipoprotein assembly and secretion in bovine hepatocytes in vitro. IUBMB Life 2020, 72, 2710–2721. [Google Scholar] [CrossRef]
- Ogata, M.; Hino, S.I.; Saito, A.; Morikawa, K.; Kondo, S.; Kanemoto, S.; Murakami, T.; Taniguchi, M.; Tanii, I.; Yoshinaga, K.; et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 2006, 26, 9220–9231. [Google Scholar] [CrossRef] [Green Version]
- Ganley, I.G.; Wong, P.M.; Gammoh, N.; Jiang, X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell. 2011, 42, 731–743. [Google Scholar] [CrossRef] [Green Version]
- Miyagawa, K.; Oe, S.; Honma, Y.; Izumi, H.; Baba, R.; Harada, M. Lipid-Induced endoplasmic reticulum stress impairs selective autophagy at the step of autophagosome-lysosome fusion in hepatocytes. Am. J. Pathol. 2016, 186, 1861–1873. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wen, J.; Kong, Y.; Zhao, C.; Liu, S.; Liu, Y.; Li, L.; Yang, J.; Zhu, X.; Zhao, B.; et al. Oxidative status in dairy goats: Periparturient variation and changes in subclinical hyperketonemia and hypocalcemia. BMC Vet Res. 2021, 17, 238. [Google Scholar] [CrossRef]
- Jump, D.B. Fatty acid regulation of gene transcription. Crit. Rev. Clin. Lab. Sci. 2004, 41, 41–78. [Google Scholar] [CrossRef]
- Amir, M.; Czaja, M.J. Autophagy in nonalcoholic steatohepatitis. Expert. Rev. Gastroenterol. Hepatol. 2011, 5, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.L.; Cuervo, A.M. Liver autophagy: Much more than just taking out the trash. Nat. Rev. Gastro. Hepat. 2014, 11, 187–200. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Alva, A.; Su, H.; Dutt, P.; Freundt, E.; Welsh, S.; Baehrecke, E.H.; Lenardo, M.J. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004, 304, 1500–1502. [Google Scholar] [CrossRef]
- Trachsel-Moncho, L.; Benlloch-Navarro, S.; Fernandez-Carbonell, A.; Ramirez-Lamelas, D.T.; Olivar, T.; Silvestre, D.; Poch, E.; Miranda, M. Oxidative stress and autophagy-related changes during retinal degeneration and development. Cell Death Dis. 2018, 9, 812. [Google Scholar] [CrossRef] [Green Version]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homolog of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2003, 22, 4577. [Google Scholar]
- Xiao, J.; Zhu, X.; He, B.; Zhang, Y.; Kang, B.; Wang, Z.; Ni, X. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J. Biomed. Sci. 2011, 18, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Li, G.; Du, X.; Sun, X.; Peng, Z.; Zhao, C.; Xu, Q.; Abdelatty, A.M.; Mohamed, F.F.; Wang, Z.; et al. Increased autophagy mediates the adaptive mechanism of the mammary gland in dairy cows with hyperketonemia. J. Dairy Sci. 2020, 103, 2545–2555. [Google Scholar] [CrossRef] [PubMed]
- Kouroku, Y.; Fujita, E.; Tanida, I.; Ueno, T.; Isoai, A.; Kumagai, H.; Ogawa, S.; Kaufman, R.J.; Kominami, E.; Momoi, T. ER stress (PERK/eIF2 alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007, 14, 230–239. [Google Scholar] [CrossRef] [Green Version]
- B’chir, W.; Maurin, A.C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2 alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic. Acids. Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.S.; Lee, H.M.; Kim, J.K.; Yang, C.S.; Kim, T.S.; Jung, M.; Jin, H.S.; Kim, S.; Jang, J.; Oh, G.T.; et al. PPAR-alpha activation mediates innate host defense through induction of tfeb and lipid catabolism. J. Immunol. 2017, 198, 3283–3295. [Google Scholar] [CrossRef] [Green Version]
- Qiu, L.; Wu, X.; Chau, J.F.; Szeto, I.Y.; Tam, W.Y.; Guo, Z.; Chung, S.K.; Oates, P.J.; Chung, S.S.; Yang, J.Y. Aldose reductase regulates hepatic peroxisome proliferator-activated receptor alpha phosphorylation and activity to impact lipid homeostasis. J. Biol. Chem. 2008, 283, 17175–17183. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Wagner, M.; Xiao, R.; Kim, K.H.; Feng, D.; Lazar, M.A.; Moore, D.D. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014, 516, 112–115. [Google Scholar] [CrossRef]
- Liu, K.; Czaja, M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.L.; Liu, G.W.; Loor, J.J.; Fang, Z.Y.; Bucktrout, R.; Yang, Y.C.; Ye, Q.Q.; Shi, Z.; Shen, T.Y.; Wang, X.H.; et al. Impaired hepatic autophagic activity in dairy cows with severe fatty liver is associated with inflammation and reduced liver function. J. Dairy Sci. 2018, 101, 11175–11185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, W.X.; Li, M.; Chen, X.; Ni, H.M.; Lin, C.W.; Gao, W.; Lu, B.; Stolz, D.B.; Clemens, D.L.; Yin, X.M. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 2010, 139, 1740–1752. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Molusky, M.M.; Song, J.; Hu, C.R.; Fang, F.; Rui, C.; Mathew, A.V.; Pennathur, S.; Liu, F.; Cheng, J.X.; et al. Autophagy deficiency by hepatic FIP200 deletion uncouples steatosis from liver injury in NAFLD. Mol. Endocrinol. 2013, 27, 1643–1654. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Zhao, C.; Liu, Y.; Kong, Y.; Tan, P.; Liu, S.; Zeng, F.; Yuan, Y.; Li, X.; Liu, G.; et al. NEFA Promotes Autophagosome Formation through Modulating PERK Signaling Pathway in Bovine Hepatocytes. Animals 2021, 11, 3400. https://doi.org/10.3390/ani11123400
Huang Y, Zhao C, Liu Y, Kong Y, Tan P, Liu S, Zeng F, Yuan Y, Li X, Liu G, et al. NEFA Promotes Autophagosome Formation through Modulating PERK Signaling Pathway in Bovine Hepatocytes. Animals. 2021; 11(12):3400. https://doi.org/10.3390/ani11123400
Chicago/Turabian StyleHuang, Yan, Chenxu Zhao, Yaoquan Liu, Yezi Kong, Panpan Tan, Siqi Liu, Fangyuan Zeng, Yang Yuan, Xinwei Li, Guowen Liu, and et al. 2021. "NEFA Promotes Autophagosome Formation through Modulating PERK Signaling Pathway in Bovine Hepatocytes" Animals 11, no. 12: 3400. https://doi.org/10.3390/ani11123400
APA StyleHuang, Y., Zhao, C., Liu, Y., Kong, Y., Tan, P., Liu, S., Zeng, F., Yuan, Y., Li, X., Liu, G., Zhao, B., & Wang, J. (2021). NEFA Promotes Autophagosome Formation through Modulating PERK Signaling Pathway in Bovine Hepatocytes. Animals, 11(12), 3400. https://doi.org/10.3390/ani11123400







