Genome-Wide Identification of Reference Genes for Reverse-Transcription Quantitative PCR in Goat Rumen
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, RNA Isolation, and cDNA Synthesis
2.2. Genome-Wide Selection of the Candidate RGs
2.3. Quantitative Real-Time PCR (RT-qPCR) and Amplification Efficiency
2.4. The Expression Stability of the Candidate RGs in Rumens
2.5. Statistical Analyses
3. Results
3.1. The Selection of RGs in Goat Rumen Tissues
3.2. RNA Purity, Primer Verification, and Amplification Efficiency
3.3. Gene Expression Dispersion Analysis
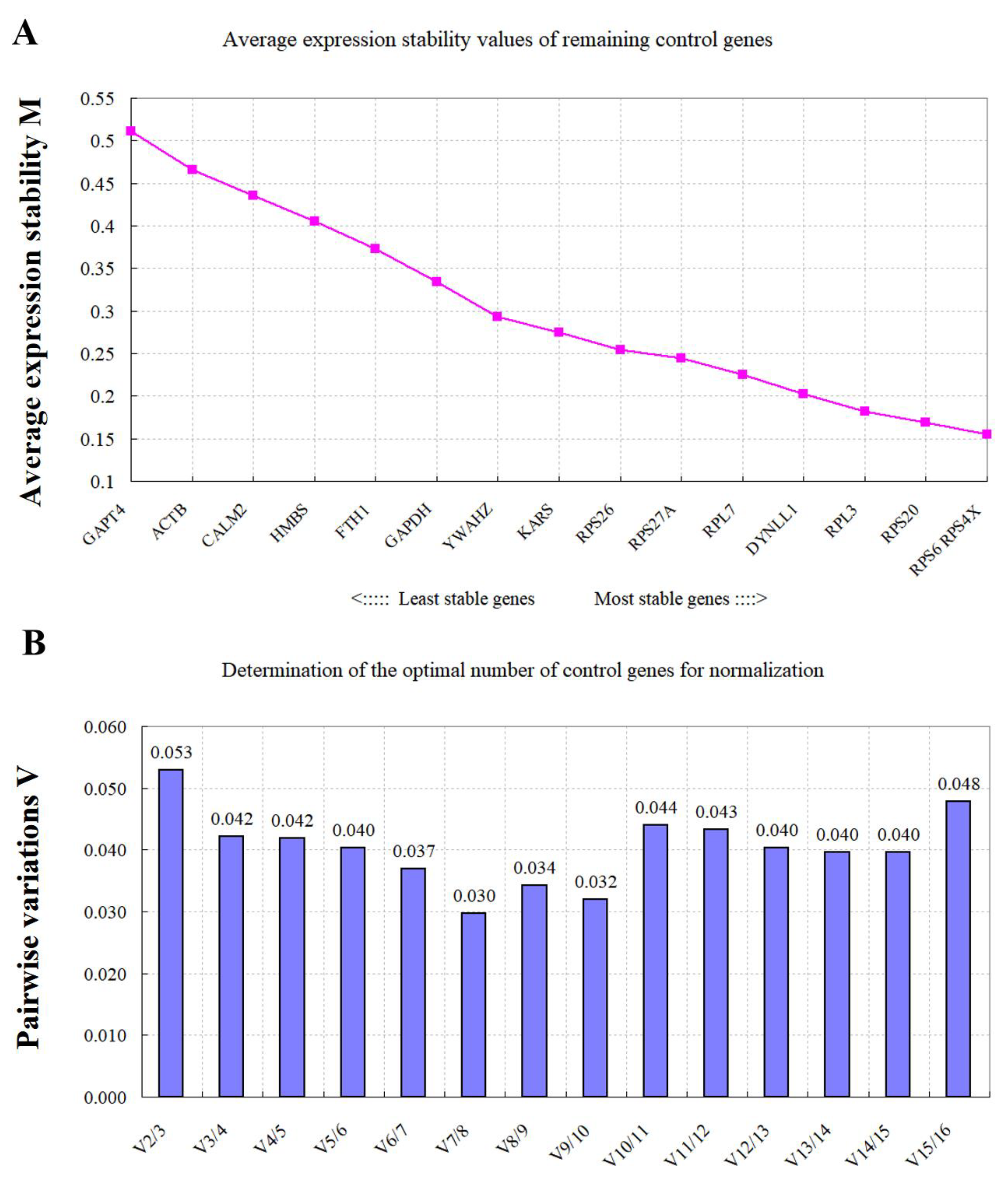
3.4. Expression Stability of the RGs Assessed via geNorm Analysis
3.5. Expression Stability of the RGs Assessed via NormFinder Analysis
3.6. Expression Stability of the RGs Assessed via Bestkeeper Analysis
3.7. Normalizing the Expression Profiles of Target Genes Using the Target RGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piazza, V.G.; Bartke, A.; Miquet, J.G.; Sotelo, A.I. Analysis of Different Approaches for the Selection of Reference Genes in RT-qPCR Experiments: A Case Study in Skeletal Muscle of Growing Mice. Int. J. Mol. Sci. 2017, 18, 1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 0034.1–0034.11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakeel, M.; Rodriguez, A.; Tahir, U.B.; Jin, F. Gene expression studies of reference genes for quantitative real-time PCR: An overview in insects. Biotechnol. Lett. 2018, 40, 227–236. [Google Scholar] [CrossRef]
- Zárybnický, T.; Matoušková, P.; Ambrož, M.; Šubrt, Z.; Skálová, L.; Boušová, I. The Selection and Validation of Reference Genes for mRNA and microRNA Expression Studies in Human Liver Slices Using RT-qPCR. Genes 2019, 10, 763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.; Wu, Z.; Cai, D.; Lu, B. Evaluation of reference genes for gene expression studies in mouse and N2a cell ischemic stroke models using quantitative real-time PCR. BMC Neurosci. 2018, 19, 3. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Dunislawska, A.; Slawinska, A.; Siwek, M. Validation of the Reference Genes for the Gene Expression Studies in Chicken DT40 Cell Line. Genes 2020, 11, 372. [Google Scholar] [CrossRef] [Green Version]
- Smits, K.; Goossens, K.; Van Soom, A.; Govaere, J.; Hoogewijs, M.; Vanhaesebrouck, E.; Galli, C.; Colleoni, S.; Vandesompele, J.; Peelman, L. Selection of reference genes for quantitative real-time PCR in equine in vivo and fresh and frozen-thawed in vitro blastocysts. BMC Res. Notes 2009, 2, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, P.D.; Sladek, R.; Greenwood, C.M.; Hudson, T.J. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002, 12, 292–297. [Google Scholar] [CrossRef] [Green Version]
- Panina, Y.; Germond, A.; Masui, S.; Watanabe, T.M. Validation of Common Housekeeping Genes as Reference for qPCR Gene Expression Analysis During iPS Reprogramming Process. Sci. Rep. 2018, 8, 8716. [Google Scholar] [CrossRef] [PubMed]
- Abuna, R.P.F.; Oliveira, F.S.; Ramos, J.I.R.; Lopes, H.B.; Freitas, G.P.; Souza, A.T.P.; Beloti, M.M.; Rosa, A.L. Selection of reference genes for quantitative real-time polymerase chain reaction studies in rat osteoblasts. J. Cell Physiol. 2018, 234, 749–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jonge, H.J.; Fehrmann, R.S.; de Bont, E.S.; Hofstra, R.M.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.; van der Zee, A.G.; te Meerman, G.J.; ter Elst, A. Evidence based selection of housekeeping genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Chen, Y.; Zhang, X.; Bai, B.; Yan, H.; Qin, D.; Xia, Q. Selection of reference genes for tissue/organ samples on day 3 fifth-instar larvae in silkworm, Bombyx mori. Arch. Insect. Biochem. Physiol. 2018, 98, e21458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Y.Q.; Jin, G.; Wu, S.; Cui, J.; Wang, R.F. Selection of reference genes for gene expression studies in human bladder cancer using SYBR-Green quantitative polymerase chain reaction. Oncol. Lett. 2017, 14, 6001–6011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, T.V.C.; Oliveira, R.L.; Menezes, D.R.; de Lucena, A.R.F.; Queiroz, M.; Lima, A.; Ribeiro, R.D.X.; Bezerra, L.R. Effects of condensed tannin-amended cassava silage blend diets on feeding behavior, digestibility, nitrogen balance, milk yield and milk composition in dairy goats. Animal 2021, 15, 100015. [Google Scholar] [CrossRef]
- Bowen, J.M.; Cormican, P.; Lister, S.J.; McCabe, M.S.; Duthie, C.A.; Roehe, R.; Dewhurst, R.J. Links between the rumen microbiota, methane emissions and feed efficiency of finishing steers offered dietary lipid and nitrate supplementation. PLoS ONE 2020, 15, e0231759. [Google Scholar] [CrossRef]
- Hu, Y.; He, Y.; Gao, S.; Liao, Z.; Lai, T.; Zhou, H.; Chen, Q.; Li, L.; Gao, H.; Lu, W. The effect of a diet based on rice straw co-fermented with probiotics and enzymes versus a fresh corn Stover-based diet on the rumen bacterial community and metabolites of beef cattle. Sci. Rep. 2020, 10, 10721. [Google Scholar] [CrossRef]
- Stumpff, F.; Martens, H.; Bilk, S.; Aschenbach, J.R.; Gäbel, G. Cultured ruminal epithelial cells express a large-conductance channel permeable to chloride, bicarbonate, and acetate. Pflug. Arch. 2009, 457, 1003–1022. [Google Scholar] [CrossRef] [Green Version]
- Yohe, T.T.; Schramm, H.; White, R.R.; Hanigan, M.D.; Parsons, C.L.M.; Tucker, H.L.M.; Enger, B.D.; Hardy, N.R.; Daniels, K.M. Form of calf diet and the rumen. II: Impact on volatile fatty acid absorption. J. Dairy Sci. 2019, 102, 8502–8512. [Google Scholar] [CrossRef] [PubMed]
- Kramer, T.; Michelberger, T.; Gürtler, H.; Gäbel, G. Absorption of short-chain fatty acids across ruminal epithelium of sheep. J. Comp. Physiol. B 1996, 166, 262–269. [Google Scholar] [CrossRef]
- Müller, F.; Huber, K.; Pfannkuche, H.; Aschenbach, J.R.; Breves, G.; Gäbel, G. Transport of ketone bodies and lactate in the sheep ruminal epithelium by monocarboxylate transporter 1. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, G1139–G1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, T.; Wang, C.; Hu, J.; Chen, X.; Niu, L.; Zhan, S.; Wang, L.; Guo, J.; Cao, J.; Li, L.; et al. Comparison of MicroRNA Transcriptomes Reveals the Association between MiR-148a-3p Expression and Rumen Development in Goats. Animals 2020, 10, 1951. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, K.; Suzuki, Y.; Roh, S. Ruminal epithelial insulin-like growth factor-binding proteins 2, 3, and 6 are associated with epithelial cell proliferation. Anim. Sci. J. 2020, 91, e13422. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019, 20, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, D.N.; Dudemaine, P.L.; Fomenky, B.E.; Ibeagha-Awemu, E.M. Integration of miRNA weighted gene co-expression network and miRNA-mRNA co-expression analyses reveals potential regulatory functions of miRNAs in calf rumen development. Genomics 2019, 111, 849–859. [Google Scholar] [CrossRef]
- Zhong, T.; Hu, J.; Xiao, P.; Zhan, S.; Wang, L.; Guo, J.; Li, L.; Zhang, H.; Niu, L. Identification and Characterization of MicroRNAs in the Goat (Capra hircus) Rumen during Embryonic Development. Front. Genet. 2017, 8, 163. [Google Scholar] [CrossRef] [Green Version]
- Die, J.V.; Baldwin, R.L.; Rowland, L.J.; Li, R.; Oh, S.; Li, C.; Connor, E.E.; Ranilla, M.J. Selection of internal reference genes for normalization of reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis in the rumen epithelium. PLoS ONE 2017, 12, e0172674. [Google Scholar] [CrossRef]
- Zhang, J.; Deng, C.; Li, J.; Zhao, Y. Transcriptome-based selection and validation of optimal house-keeping genes for skin research in goats (Capra hircus). BMC Genom. 2020, 21, 493. [Google Scholar] [CrossRef]
- China National Commission of Animal Genetic Resources. Animal Genetic Resources in China: Sheep and Goats; China Agricultural Press: Beijing, China, 2011; pp. 339–342. [Google Scholar]
- Li, D.; Hu, B.; Wang, Q.; Liu, H.; Pan, F.; Wu, W. Identification and Evaluation of Reference Genes for Accurate Transcription Normalization in Safflower under Different Experimental Conditions. PLoS ONE 2015, 10, e0140218. [Google Scholar] [CrossRef]
- Dhayat, S.A.; Abdeen, B.; Köhler, G.; Senninger, N.; Haier, J.; Mardin, W.A. MicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage II. Clin. Epigenetics 2015, 7, 132. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Cocci, P.; Mosconi, G.; Palermo, F.A. Changes in expression of microRNA potentially targeting key regulators of lipid metabolism in primary gilthead sea bream hepatocytes exposed to phthalates or flame retardants. Aquat. Toxicol. 2019, 209, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, X.; Ding, X.; Chu, M.; Liang, C.; Pei, J.; Xiong, L.; Bao, P.; Guo, X.; Yan, P. Reference gene selection and myosin heavy chain (MyHC) isoform expression in muscle tissues of domestic yak (Bos grunniens). PLoS ONE 2020, 15, e0228493. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Lin, Y.; Liao, H.; Wang, Y. Selection of reference genes for gene expression studies related to intramuscular fat deposition in Capra hircus skeletal muscle. PLoS ONE 2015, 10, e0121280. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, K.; Chen, F.; Li, W.; Sun, S.; Shi, X.E.; Yang, G. Verification of suitable and reliable reference genes for quantitative real-time PCR during adipogenic differentiation in porcine intramuscular stromal-vascular cells. Animal 2016, 10, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Najafpanah, M.J.; Sadeghi, M.; Bakhtiarizadeh, M.R. Reference genes selection for quantitative real-time PCR using RankAggreg method in different tissues of Capra hircus. PLoS ONE 2013, 8, e83041. [Google Scholar] [CrossRef]
- Tsotetsi, T.N.; Collins, N.E.; Oosthuizen, M.C.; Sibeko-Matjila, K.P. Selection and evaluation of housekeeping genes as endogenous controls for quantification of mRNA transcripts in Theileria parva using quantitative real-time polymerase chain reaction (qPCR). PLoS ONE 2018, 13, e0196715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezera, M.A.; Li, W.; Edwards, A.J.; Koch, D.J.; Beard, A.D.; Wiltbank, M.C. Identification of stable genes in the corpus luteum of lactating Holstein cows in pregnancy and luteolysis: Implications for selection of reverse-transcription quantitative PCR reference genes. J. Dairy Sci. 2020, 103, 4846–4857. [Google Scholar] [CrossRef]
- Pan, X.; Li, Z.; Li, B.; Zhao, C.; Wang, Y.; Chen, Y.; Jiang, Y. Dynamics of rumen gene expression, microbiome colonization, and their interplay in goats. BMC Genom. 2021, 22, 288. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Liu, X.; Yan, J.; Aabdin, Z.U.; Bilal, M.S.; Shen, X. Sodium Butyrate Ameliorates High-Concentrate Diet-Induced Inflammation in the Rumen Epithelium of Dairy Goats. J. Agric. Food Chem. 2017, 65, 596–604. [Google Scholar] [CrossRef]
- Abecia, L.; Jiménez, E.; Martínez-Fernandez, G.; Martín-García, A.I.; Ramos-Morales, E.; Pinloche, E.; Denman, S.E.; Newbold, C.J.; Yáñez-Ruiz, D.R. Natural and artificial feeding management before weaning promote different rumen microbial colonization but not differences in gene expression levels at the rumen epithelium of newborn goats. PLoS ONE 2017, 12, e0182235. [Google Scholar] [CrossRef] [PubMed]
- Lechuga, C.G.; Simón-Carrasco, L.; Jacob, H.K.; Drosten, M. Genetic Validation of Cell Proliferation via Ras-Independent Activation of the Raf/Mek/Erk Pathway. Methods Mol. Biol. 2017, 1487, 269–276. [Google Scholar] [CrossRef]
- Mattox, T.E.; Chen, X.; Maxuitenko, Y.Y.; Keeton, A.B.; Piazza, G.A. Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers. Int. J. Mol. Sci. 2019, 21, 141. [Google Scholar] [CrossRef] [Green Version]
- Ribó, P.; Guo, Y.; Aranda, J.; Ainsua-Enrich, E.; Navinés-Ferrer, A.; Guerrero, M.; Pascal, M.; de la Cruz, C.; Orozco, M.; Muñoz-Cano, R.; et al. Mutation in KARS: A novel mechanism for severe anaphylaxis. J. Allergy Clin. Immunol. 2021, 147, 1855–1864.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, J.B.; Zeng, Q.Y.; Wu, S.; Xue, M.Q.; Fang, P.; Wang, E.D.; Zhou, X.L. Hearing impairment-associated KARS mutations lead to defects in aminoacylation of both cytoplasmic and mitochondrial tRNA(Lys). Sci. China Life Sci. 2020, 63, 1227–1239. [Google Scholar] [CrossRef]
- Macabelli, C.H.; Ferreira, R.M.; Gimenes, L.U.; de Carvalho, N.A.; Soares, J.G.; Ayres, H.; Ferraz, M.L.; Watanabe, Y.F.; Watanabe, O.Y.; Sangalli, J.R.; et al. Reference gene selection for gene expression analysis of oocytes collected from dairy cattle and buffaloes during winter and summer. PLoS ONE 2014, 9, e93287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Petibon, C.; Malik, G.M.; Catala, M.; Abou, E.S. Regulation of ribosomal protein genes: An ordered anarchy. Wiley Interdiscip. Rev. RNA 2021, 12, e1632. [Google Scholar] [CrossRef]
- Daftuar, L.; Zhu, Y.; Jacq, X.; Prives, C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE 2013, 8, e68667. [Google Scholar] [CrossRef]
- Xiong, X.; Zhao, Y.; He, H.; Sun, Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene 2011, 30, 1798–1811. [Google Scholar] [CrossRef] [Green Version]
- Goudarzi, K.M.; Lindström, M.S. Role of ribosomal protein mutations in tumor development (Review). Int. J. Oncol. 2016, 48, 1313–1324. [Google Scholar] [CrossRef] [Green Version]
- Khalaileh, A.; Dreazen, A.; Khatib, A.; Apel, R.; Swisa, A.; Kidess-Bassir, N.; Maitra, A.; Meyuhas, O.; Dor, Y.; Zamir, G. Phosphorylation of ribosomal protein S6 attenuates DNA damage and tumor suppression during development of pancreatic cancer. Cancer Res. 2013, 73, 1811–1820. [Google Scholar] [CrossRef] [Green Version]
- Sudhamalla, B.; Kumar, M.; Roy, K.R.; Kumar, R.S.; Bhuyan, A.K. Cysteine endoprotease activity of human ribosomal protein S4 is entirely due to the C-terminal domain, and is consistent with Michaelis-Menten mechanism. Biochim. Biophys. Acta 2013, 1830, 5342–5349. [Google Scholar] [CrossRef]
- Zinn, A.R.; Alagappan, R.K.; Brown, L.G.; Wool, I.; Page, D.C. Structure and function of ribosomal protein S4 genes on the human and mouse sex chromosomes. Mol. Cell Biol. 1994, 14, 2485–2492. [Google Scholar] [CrossRef] [Green Version]



| Gene | Accession No. | Sequences (5′-3′) | Tm (°C) | Size (bp) | Slope | Efficiency (%) | R2 | |
|---|---|---|---|---|---|---|---|---|
| RPS20 | XM_013969227.2 | F: ATCAGAGGCGCGAAGGAAAA | 56.9 | 158 | −3.421 | 96.0% | 1.000 | |
| R: TGCAGGTCAATGAGTCGCTT | ||||||||
| RPL7 | XM_005689063.3 | F: ACTTCCTGTGGCCCTTTAA | 56.9 | 103 | −3.489 | 93.5% | 0.993 | |
| R: ATCTGGTCTTCCCTGTTGC | ||||||||
| RPL3 | XM_005681086.3 | F: CTGACAAGAGCATCAACCC | 56.9 | 209 | −3.472 | 94.1% | 0.999 | |
| R: GAAGCGACCATGACCAAAT | ||||||||
| RPS26 | XM_013963957.2 | F: GAACAACGGTCGTGCCAAAA | 56.9 | 171 | −3.431 | 95.6% | 0.993 | |
| R: ACGTAGGCGTCGAAAACACT | ||||||||
| RPS4X | XM_005700650.3 | F: TACTTGGCCTCCTCAGGTGT | 59.4 | 223 | −3.178 | 106.4% | 0.999 | |
| R: TACTTGGCCTCCTCAGGTGT | ||||||||
| RPS27A | XM_005686612.3 | F: TCTAGTGTTGAGACTTCGTGGTG | 59.4 | 183 | −3.523 | 92.3% | 0.997 | |
| R: CCAGCACCACATTCATCTGAGG | ||||||||
| GAPDH | XM_005680968.3 | F: GCAAGTTCCACGGCACAG | 59.4 | 249 | −3.398 | 96.9% | 1.000 | |
| R: GGTTCACGCCCATCACAA | ||||||||
| CALM2 | XM_005686574.3 | F: AGAAGCATTCCGTGTGTTT | 56.9 | 159 | −3.495 | 93.3% | 0.995 | |
| R: TCATAGTTTACTTGACCAT | ||||||||
| RPS6 | XM_005683632.3 | F: GGACTGGAGAGAGAAAGCG | 59.4 | 211 | −3.324 | 99.9% | 0.996 | |
| R: ACAACATACTGGCGGACAT | ||||||||
| FTH1 | NM_001285609.1 | F: GCTTGGAAAGAAGTGTGAA | 56.9 | 153 | −3.364 | 98.3% | 0.992 | |
| R: GCAGGTTGGTTATGTGGTC | ||||||||
| DYNLL1 | XM_018061128.1 | F: GCCGTAATCAAGAATGCCGA | 56.9 | 172 | −3.285 | 101.6% | 1.000 | |
| R: CGAAGTTCCTCCCCACGATG | ||||||||
| KARS | XM_005691813.3 | F: AATCACAGTGCTGATGATGGCA | 59.4 | 94 | −3.256 | 102.8% | 0.999 | |
| R: TCAGCTGGTGGATTGCTTGG | ||||||||
| ACTB | XM_018039831.1 | F: CCTGCGGCATTCACGAAACTAC | 59.4 | 87 | −3.223 | 104.3% | 0.997 | |
| R: ACAGCACCGTGTTGGCGTAGAG | ||||||||
| YWHAZ | XM_018058314.1 | F: ACTACTATCGCTACTTGGCTGAG | 59.4 | 84 | −3.264 | 102.5% | 0.998 | |
| R: CTTCTTGTTATGCTTGCTGTGA | ||||||||
| GPAT4 | XM_018041983.1 | F: GGAGTCTCCTTTGGTATCCG | 56.9 | 128 | −3.165 | 107.0% | 0.992 | |
| R: CCATTGGTGTAGGGCTTGTA | ||||||||
| HMBS | XM_005689536.3 | F: GCAACGGCGGAAGAAGACA | 59.4 | 267 | −3.316 | 100.3% | 0.994 | |
| R: CAGCGAGTGAACAACCAGG | ||||||||
| TOP2A | XM_005693780.3 | F: AGCCCATTGGTCAGTTTGGT | 55.0 | 218 | - | - | - | |
| R: ACCAATTCCTTCAGCGCCAT | ||||||||
| IGF1 | XM_005680537.3 | F: CAGTCACATCCTCCTCGCAT | 61.3 | 112 | - | - | - | |
| R: AGAGCATCCACCAACTCAGC |
| Gene Symbol | SD | CV | r | Rank Order |
|---|---|---|---|---|
| RPS4X | 0.33 | 1.79 | 0.973 | 1 |
| RPS6 | 0.38 | 2.15 | 0.943 | 2 |
| DYNLL1 | 0.36 | 1.71 | 0.930 | 3 |
| RPL7 | 0.31 | 1.75 | 0.917 | 4 |
| RPS20 | 0.36 | 2.05 | 0.916 | 5 |
| RPL3 | 0.37 | 2.03 | 0.914 | 6 |
| RPS26 | 0.35 | 1.99 | 0.889 | 7 |
| RPS27A | 0.26 | 1.49 | 0.872 | 8 |
| YWAHZ | 0.33 | 1.58 | 0.794 | 9 |
| KARS | 0.32 | 1.46 | 0.730 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Wang, C.; Zhang, L.; Lei, A.; Wang, L.; Niu, L.; Zhan, S.; Guo, J.; Cao, J.; Li, L.; et al. Genome-Wide Identification of Reference Genes for Reverse-Transcription Quantitative PCR in Goat Rumen. Animals 2021, 11, 3137. https://doi.org/10.3390/ani11113137
Zhao J, Wang C, Zhang L, Lei A, Wang L, Niu L, Zhan S, Guo J, Cao J, Li L, et al. Genome-Wide Identification of Reference Genes for Reverse-Transcription Quantitative PCR in Goat Rumen. Animals. 2021; 11(11):3137. https://doi.org/10.3390/ani11113137
Chicago/Turabian StyleZhao, Juan, Cheng Wang, Lin Zhang, Aiai Lei, Linjie Wang, Lili Niu, Siyuan Zhan, Jiazhong Guo, Jiaxue Cao, Li Li, and et al. 2021. "Genome-Wide Identification of Reference Genes for Reverse-Transcription Quantitative PCR in Goat Rumen" Animals 11, no. 11: 3137. https://doi.org/10.3390/ani11113137
APA StyleZhao, J., Wang, C., Zhang, L., Lei, A., Wang, L., Niu, L., Zhan, S., Guo, J., Cao, J., Li, L., Zhang, H., & Zhong, T. (2021). Genome-Wide Identification of Reference Genes for Reverse-Transcription Quantitative PCR in Goat Rumen. Animals, 11(11), 3137. https://doi.org/10.3390/ani11113137






