RNA-Seq Reveals the Expression Profiles of Long Non-Coding RNAs in Lactating Mammary Gland from Two Sheep Breeds with Divergent Milk Phenotype
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. RNA Extraction
2.3. Library Construction and RNA Sequencing
2.4. Sequence Analysis
2.5. Identification of lncRNA and Screening of Differentially Expressed lncRNAs
2.6. The Prediction of the Target Gene of lncRNAs and GO and KEGG Enrichment Analysis of the Target Genes
2.7. Validation of Differentially Expressed lncRNAs
2.8. Construction of the lncRNA-miRNA Network
3. Results
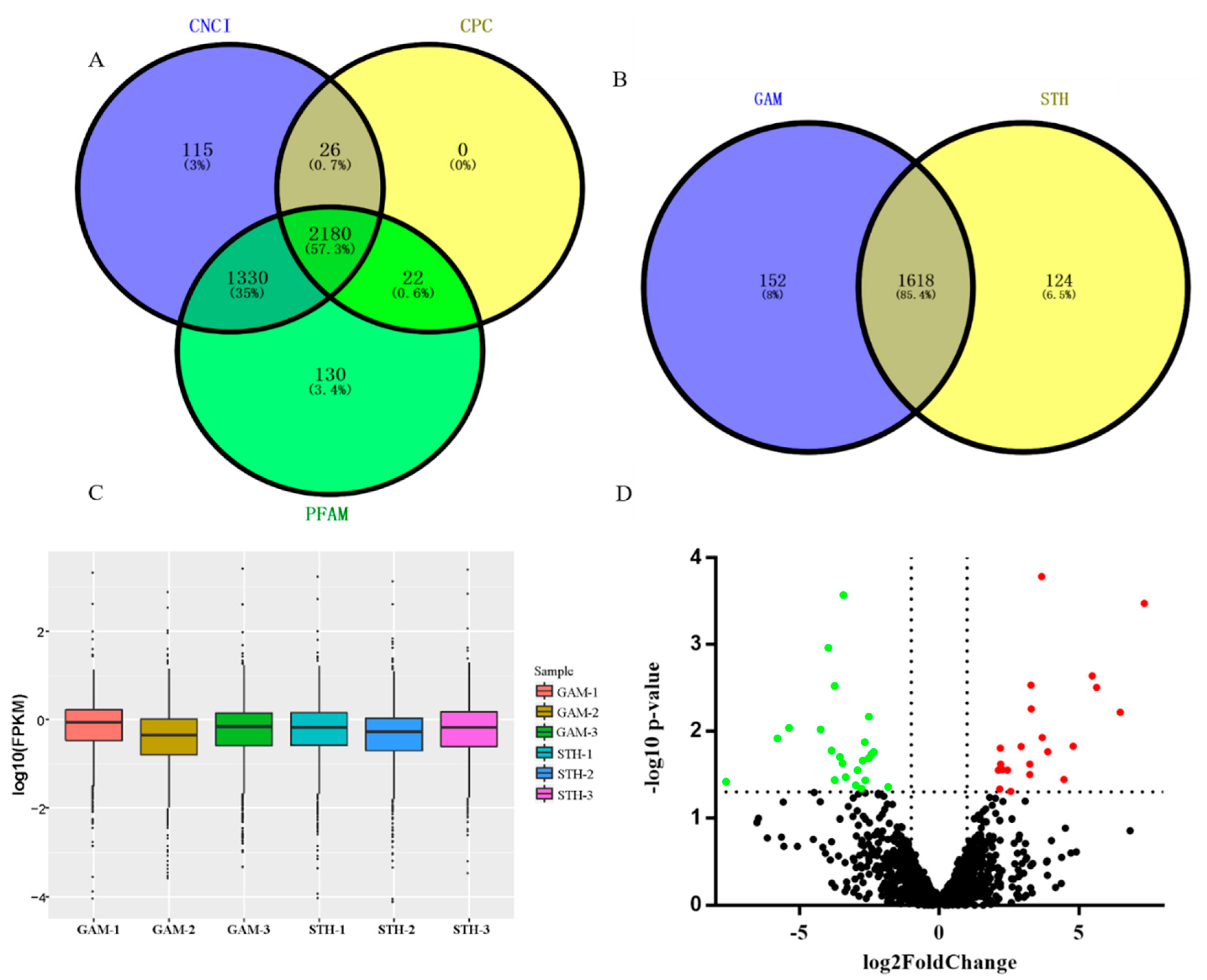
3.1. Identification of lncRNAs in the Ovine Mammary Gland
3.2. Analysis and Validation of Differentially Expressed lncRNAs
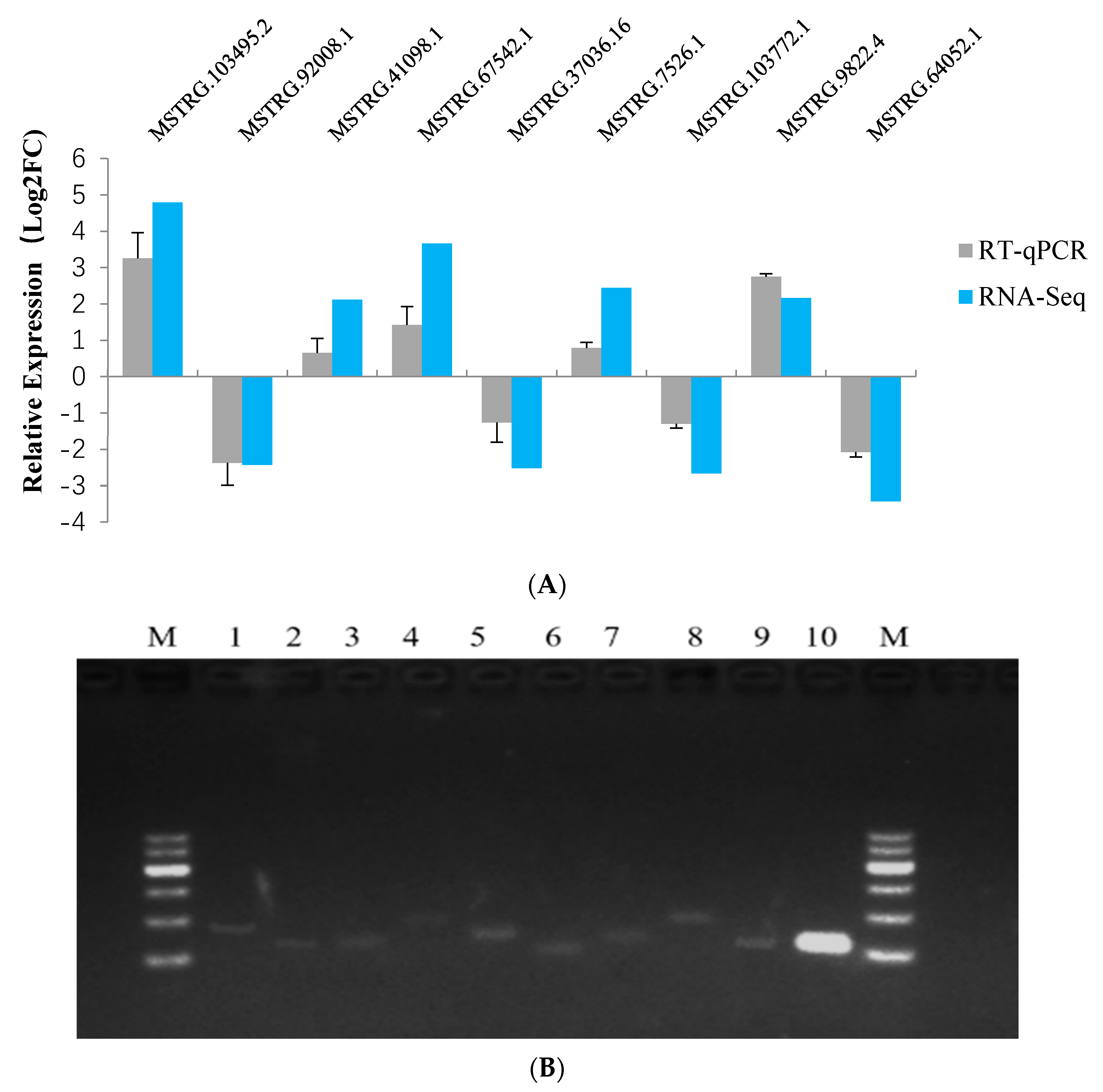
3.3. Validation of Differentially Expressed lncRNAs Using RT-qPCR
3.4. GO Enrichment and KEGG Pathway Analysis of Differentially Expressed lncRNAs
3.5. Bioinformatic Analysis of lncRNA-miRNA Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Knowling, S.; Morris, K.V. Non-coding RNA and antisense RNA. Nature’s trash or treasure? Biochimie 2011, 93, 1922–1927. [Google Scholar] [CrossRef]
- Spizzo, R.; Almeida, M.I.; Colombatti, A.; Calin, G.A. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene 2012, 31, 4577–4587. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhao, Y.; Lai, F.; Chu, M.; Hao, Y.; Feng, Y.; Zhang, H.; Liu, J.; Cheng, M.; Li, L.; et al. LncRNA as ceRNAs may be involved in lactation process. Oncotarget 2017, 8, 98014–98028. [Google Scholar] [CrossRef]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G. Long non-coding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest-and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, 8. [Google Scholar] [CrossRef]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef]
- Nishikawa, K.; Kinjo, A.R. Essential role of long non-coding RNAs in de novo chromatin modifications: The genomic address code hypothesis. Biophys. Rev. 2017, 9, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Standaert, L.; Adriaens, C.; Radaelli, E.; Van Keymeulen, A.; Blanpain, C.; Hirose, T. The long noncoding RNA neat1 is required for mammary gland development and lactation. RNA 2014, 20, 1844–1849. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Chen, Q.; Zhao, L.; Ma, J.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of long intergenic noncoding RNAs in bovine mammary glands. BMC Genomics 2017, 18, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Li, R.; Dudemaine, P.L.; Do, D.N.; Bissonnette, N. Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil. Int. J. Mol. Sci. 2018, 19, 3610. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Qiao, J.; Zhang, Z.; Shang, X.; Chu, Z.; Fu, Y.; Chu, M. Identification and analysis of differentially expressed long non-coding RNAs of Chinese Holstein cattle responses to heat stress. Anim. Biotechnol. 2020, 31, 9–16. [Google Scholar] [CrossRef]
- Yang, B.; Jiao, B.; Ge, W.; Zhang, X.; Wang, S.; Zhao, H.; Wang, X. Transcriptome sequencing to detect the potential role of long non-coding RNAs in bovine mammary gland during the dry and lactation period. BMC Genomics 2018, 19, 605. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ning, C.; Zhao, P.; Feng, W.; Jin, Y.; Zhou, L.; Yu, Y.; Liu, J. Integrated analysis of long noncoding RNA and mRNA expression profiles reveals the potential role of long noncoding RNA in different bovine lactation stages. J. Dairy Sci. 2018, 101, 11061–11073. [Google Scholar] [CrossRef]
- Ji, Z.; Chao, T.; Liu, Z.; Hou, L.; Wang, J.; Wang, A.; Zhou, J.; Xuan, R.; Wang, G.; Wang, J. Genome-wide integrated analysis demonstrates widespread functions of lncRNAs in mammary gland development and lactation in dairy goats. BMC Genomics 2020, 21, 254. [Google Scholar] [CrossRef]
- Ni, Y.; Wu, F.; Chen, Q.; Cai, J.; Hu, J.; Shen, J.; Zhang, J. Long noncoding RNA and mRNA profiling of hypothalamic-pituitary-mammary gland axis in lactating sows under heat stress. Genomics 2020, 112, 3668–3676. [Google Scholar] [CrossRef]
- Lanz, R.B.; Chua, S.S.; Barron, N.; Soder, B.M.; DeMayo, F.; O’Malley, B.W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell. Biol. 2003, 23, 7163–7176. [Google Scholar] [CrossRef]
- Ginger, M.R.; Shore, A.N.; Contreras, A.; Rijnkels, M.; Miller, J.; Gonzalez-Rimbau, M.F. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. USA 2006, 103, 5781–5786. [Google Scholar] [CrossRef] [PubMed]
- Askarian-Amiri, M.E.; Crawford, J.; French, J.D.; Smart, C.E.; Smith, M.A.; Clark, M.B. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 2011, 17, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.Y.; Zhou, H.T.; Hickford, J.G.H.; Gong, H.; Wang, J.Q.; Hu, J.; Liu, X.; Li, S.B.; Zhao, M.L.; Luo, Y.Z. Transcriptome Profile Analysis of Mammary Gland Tissue from Two Breeds of Lactating Sheep. Genes 2019, 10, 781. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.Y.; Zhou, H.T.; Hickford, J.G.H.; Gong, H.; Wang, J.Q.; Hu, J.; Liu, X.; Li, S.B.; Zhao, M.L.; Luo, Y.Z. Identification and characterization of circular RNA in lactating mammary glands from two breeds of sheep with different milk production profiles using RNA-Seq. Genomics 2020, 112, 2086–2193. [Google Scholar] [CrossRef] [PubMed]
- Paten, A.M.; Pain, S.J.; Peterson, S.W.; Blair, H.T.; Kenyon, P.R.; Dearden, P.K.; Duncan, E.J. Identification of reference genes for RT-qPCR in ovine mammary tissue during late pregnancy and lactation and in response to maternal nutritional programming. Physiol. Genomics 2014, 46, 560–570. [Google Scholar] [CrossRef]
- Paten, A.M.; Duncan, E.J.; Pain, S.J.; Peterson, S.W.; Kenyon, P.R.; Blair, H.T.; Dearden, P.K. Functional development of the adult ovine mammary gland-insights from gene expression profiling. BMC Genomics 2015, 16, 748–761. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, J.; Sun, S.; Cao, D.; Shi, H.; Loor, J.J. miR-148a and miR-17-5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biol. 2017, 14, 326–338. [Google Scholar] [CrossRef]
- Lin, X.; Luo, J.; Zhang, L.; Wang, W.; Gou, D. MiR-103 controls milk fat accumulation in goat (Capra hircus) mammary gland during lactation. PLoS ONE 2013, 8, e79258. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.Q.; Wang, Y.N.; Zan, L.S.; Yang, W.C. miR-27a controls triacylglycerol synthesis in bovine mammary epithelial cells by targeting peroxisome proliferator-activated receptor gamma. J. Dairy Sci. 2017, 100, 4102–4112. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Han, S.; Wang, Y.; Yang, Z.; Zou, Z.; Liu, J.; Zhao, Z.; Wu, R.; Wang, C. Bta-miR-152 affects intracellular triglyceride content by targeting the UCP3 gene. J. Anim. Physiol. Anim. Nutr. 2019, 103, 1365–1373. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, X.; Xie, M.; Liu, M.; Ye, M.; Li, M.; Chen, X.M.; Li, X.; Zhou, R. The NF-κB-responsive long noncoding RNA firre regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 2017, 199, 3571–3582. [Google Scholar] [CrossRef]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar]
- Desprez, P.Y.; Sumida, T.; Coppé, J.P. Helix-loop-helix proteins in mammary gland development and breast cancer. J. Mammary Gland. Biol. Neoplasia 2003, 8, 225–239. [Google Scholar] [CrossRef]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef]
- Zhao, Y.; Johansson, C.; Tran, T.; Bettencourt, R.; Itahana, Y.; Desprez, P.Y.; Konieczny, S.F. Identification of a basic helix-loop-helix transcription factor expressed in mammary gland alveolar cells and required for maintenance of the differentiated state. Mol. Endocrinol. 2006, 20, 2187–2198. [Google Scholar] [CrossRef]
- Lian, S.; Guo, J.R.; Nan, X.M.; Ma, L.; Loor, J.J.; Bu, D.P. MicroRNA Bta-miR-181a regulates the biosynthesis of bovine milk fat by targeting ACSL1. J. Dairy Sci. 2016, 99, 3916–3924. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Hagiwara, N.; Radisky, D.C.; Hirai, Y. CCAAT/enhancer binding protein beta (C/EBPβ) isoform balance as a regulator of epithelial-mesenchymal transition in mouse mammary epithelial cells. Exp. Cell Res. 2014, 327, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Manka, D.; Wagner, K.U.; Khan, S.A. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc. Natl. Acad. Sci. USA 2007, 104, 14718–14723. [Google Scholar] [CrossRef] [PubMed]
- Cros, M.; Cataisson, C.; Cho, Y.M.; Berthois, Y.; Bernard-Poenaru, O.; Denne, M.; Graulet, A.M.; De-Vernejoul, M.C.; Foley, J.; Bouizar, Z. Constitutive production of parathyroid hormone-related protein (PTHrP) by fibroblasts derived from normal and pathological human breast tissue. Oncol. Res. 2002, 13, 137–146. [Google Scholar] [PubMed]
- Wansbury, O.; Panchal, H.; James, M.; Parry, S.; Ashworth, A.; Howard, B. Dynamic expression of Erbb pathway members during early mammary gland morphogenesis. J. Investig. Dermatol. 2008, 128, 1009–1021. [Google Scholar] [CrossRef]
- Andrechek, E.R.; White, D.; Muller, W.J. Targeted disruption of ErbB2/Neu in the mammary epithelium results in impaired ductal outgrowth. Oncogene 2005, 24, 932–937. [Google Scholar] [CrossRef]
- Chu, E.Y.; Hens, J.; Andl, T.; Kairo, A.; Yamaguchi, T.P.; Brisken, C.; Glick, A.; Wysolmerski, J.J.; Millar, S.E. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development 2004, 131, 4819–4829. [Google Scholar] [CrossRef]

| KEGG Pathway/GO Term | LncRNA | Target Genes | p-Value |
|---|---|---|---|
| KEGG: ErbB signaling pathway | MSTRG.100632.1|MSTRG.87828.1 | PTK2|CAMK2D | p < 0.05 |
| KEGG: Wnt signaling pathway | MSTRG.19103.1 |MSTRG.87828.1 | CACYBP|CAMK2D | p < 0.05 |
| GO: 0061180 mammary gland epithelium development | MSTRG.23883.3|MSTRG.99147.2 |MSTRG.76074.1 | CEBPB|ESR1|PTHLH | p < 0.05 |
| GO: 0033598 mammary gland epithelial cell proliferation | MSTRG.23883.3|MSTRG.99147.2 | CEBPB|ESR1 | p < 0.05 |
| GO: 0060443 mammary gland morphogenesis | MSTRG.99147.2 |MSTRG.76074.1 | ESR1|PTHLH | p < 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Z.; Luo, Y.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Jin, X.; Ke, N.; Zhao, M.; Hu, L.; et al. RNA-Seq Reveals the Expression Profiles of Long Non-Coding RNAs in Lactating Mammary Gland from Two Sheep Breeds with Divergent Milk Phenotype. Animals 2020, 10, 1565. https://doi.org/10.3390/ani10091565
Hao Z, Luo Y, Wang J, Hu J, Liu X, Li S, Jin X, Ke N, Zhao M, Hu L, et al. RNA-Seq Reveals the Expression Profiles of Long Non-Coding RNAs in Lactating Mammary Gland from Two Sheep Breeds with Divergent Milk Phenotype. Animals. 2020; 10(9):1565. https://doi.org/10.3390/ani10091565
Chicago/Turabian StyleHao, Zhiyun, Yuzhu Luo, Jiqing Wang, Jiang Hu, Xiu Liu, Shaobin Li, Xiayang Jin, Na Ke, Mengli Zhao, Liyan Hu, and et al. 2020. "RNA-Seq Reveals the Expression Profiles of Long Non-Coding RNAs in Lactating Mammary Gland from Two Sheep Breeds with Divergent Milk Phenotype" Animals 10, no. 9: 1565. https://doi.org/10.3390/ani10091565
APA StyleHao, Z., Luo, Y., Wang, J., Hu, J., Liu, X., Li, S., Jin, X., Ke, N., Zhao, M., Hu, L., Lu, Y., Wu, X., & Qiao, L. (2020). RNA-Seq Reveals the Expression Profiles of Long Non-Coding RNAs in Lactating Mammary Gland from Two Sheep Breeds with Divergent Milk Phenotype. Animals, 10(9), 1565. https://doi.org/10.3390/ani10091565





