Analysis of lncRNA Expression Profile during the Formation of Male Germ Cells in Chickens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals
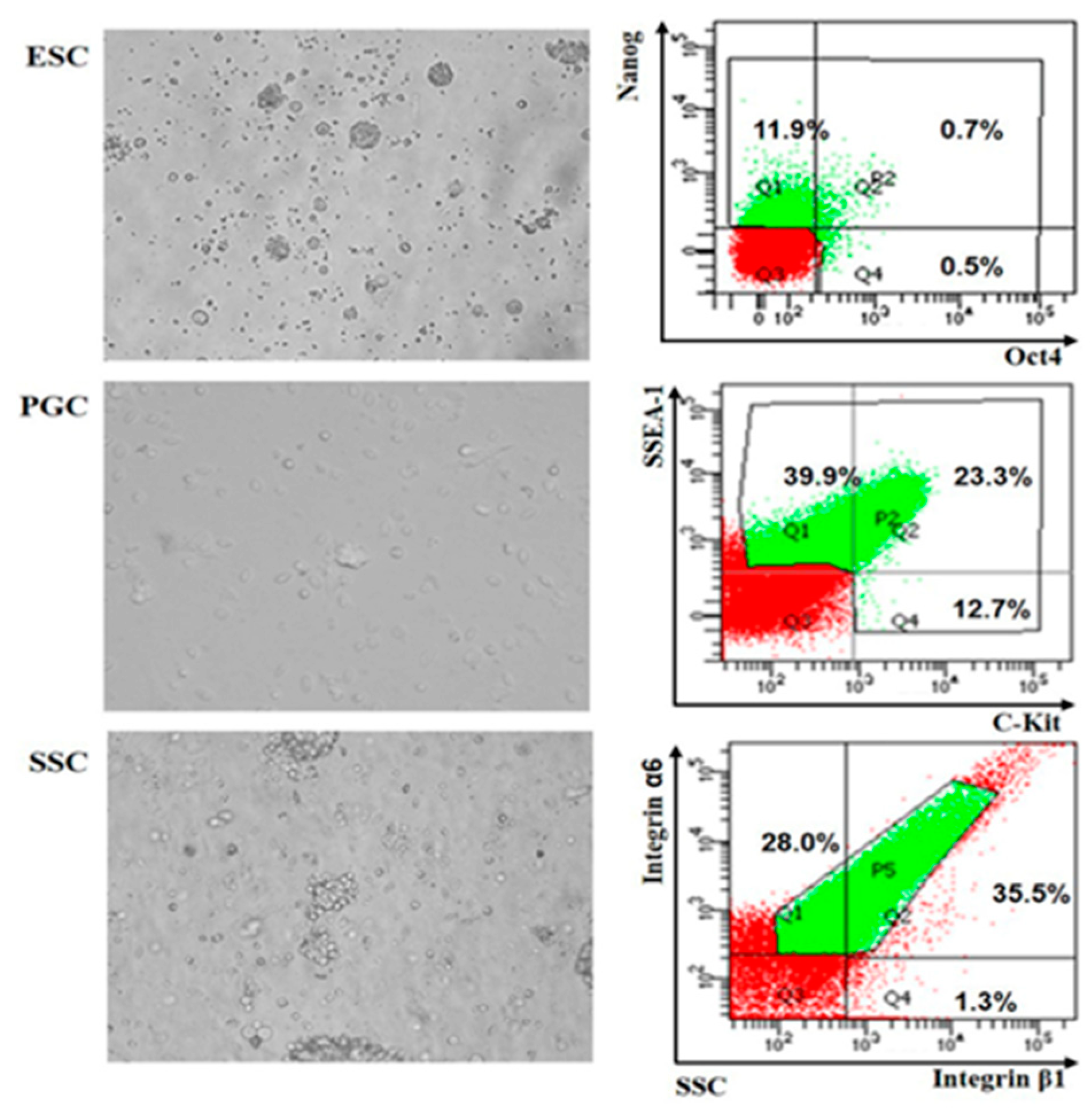
2.3. Cell Isolation and Flow Cytometry
2.4. Library Construction and Sequencing
2.5. Sequence Data Analysis
2.6. Prediction and Functional Enrichment Analysis of lncRNA Target Genes
2.7. The lncRNA–mRNA and mRNA–mRNA Co-expression Network Analysis
2.8. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Verification
3. Results
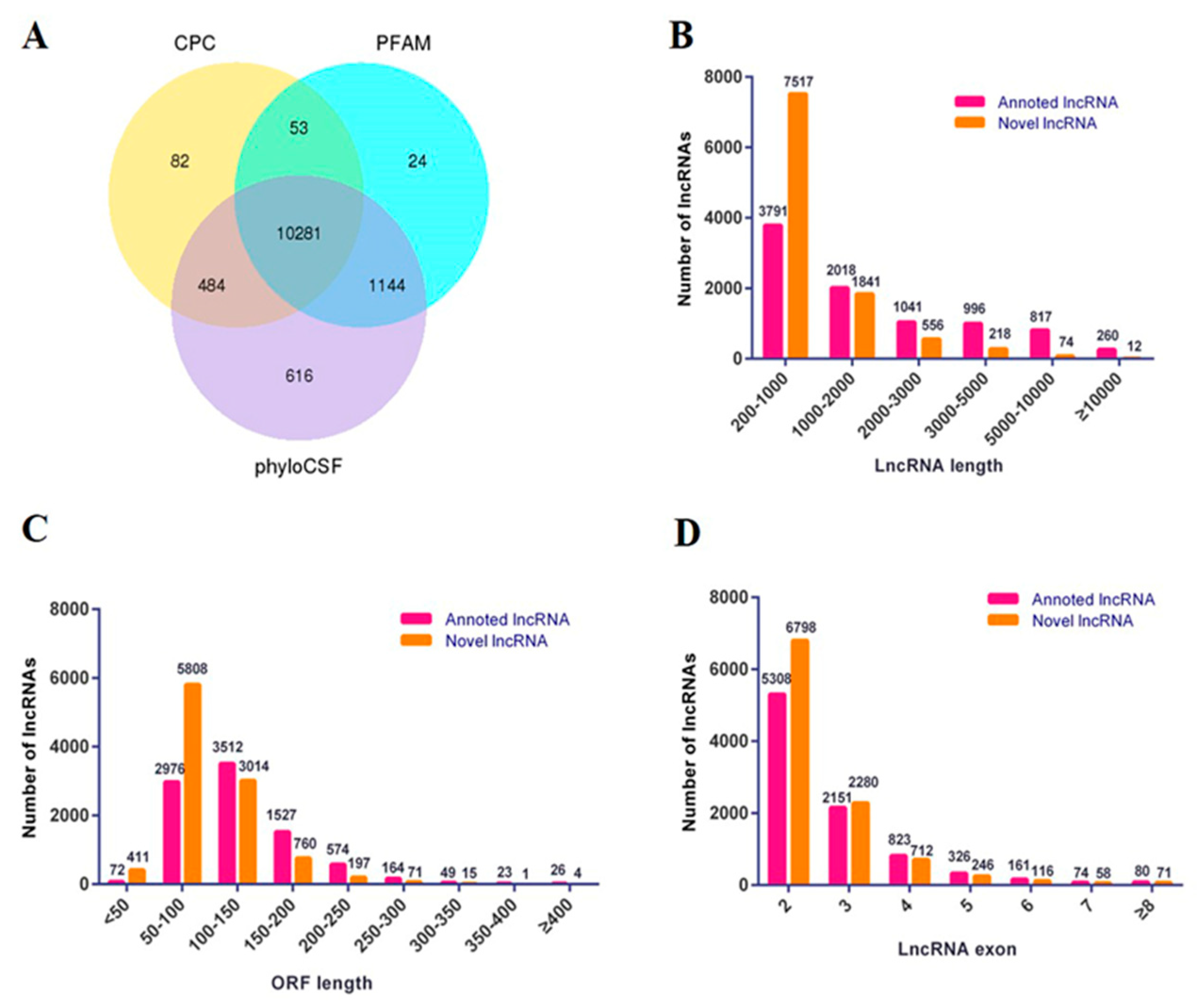
3.1. Overview of Sequencing Results
3.2. Analysis and Verification of Differentially Expressed lncRNAs in ESCs, PGCs and SSCs
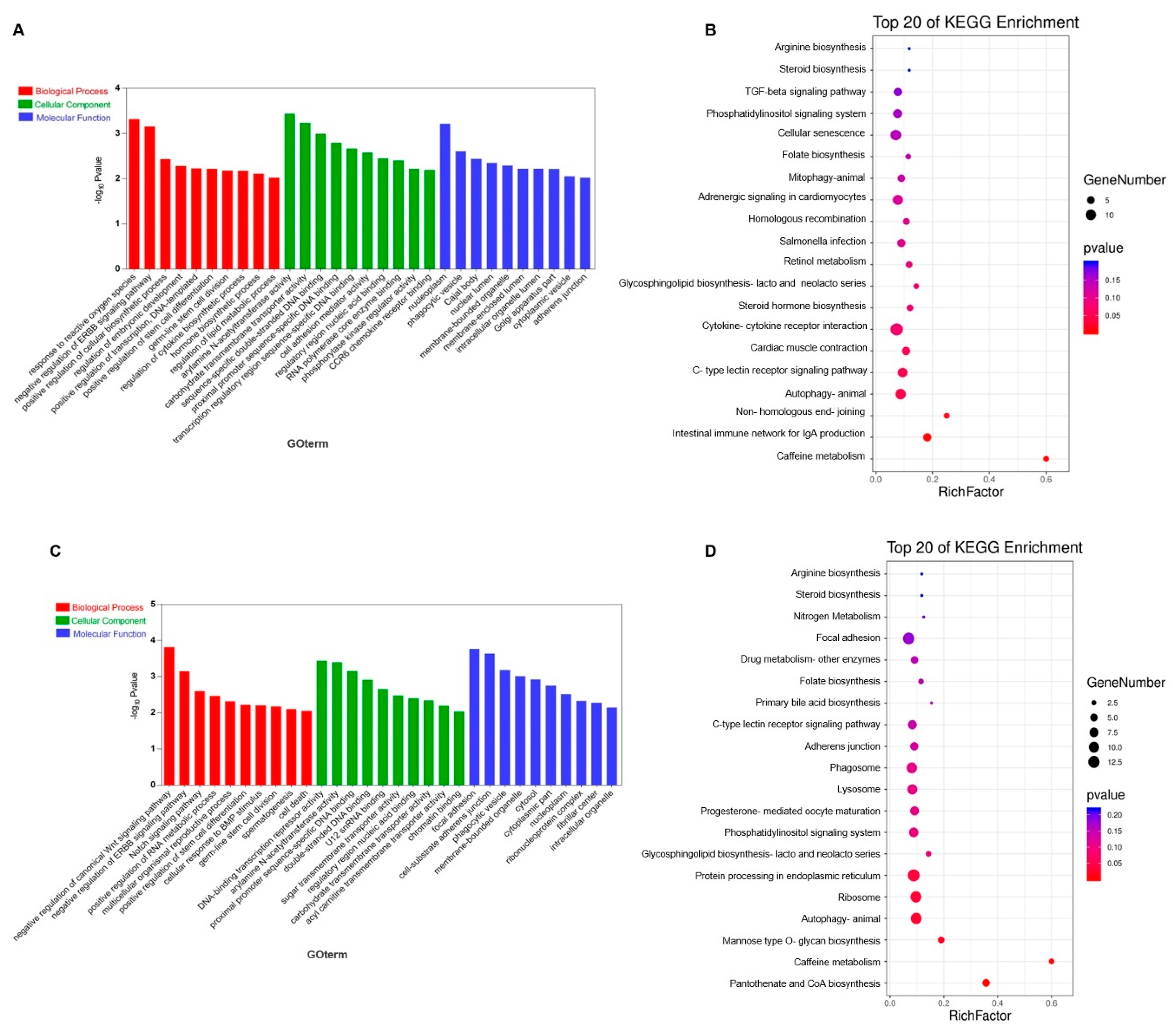
3.3. Prediction and Functional Enrichment Analysis of lncRNA Target Genes
3.4. Screening of DELs Related to Germ Cell Development
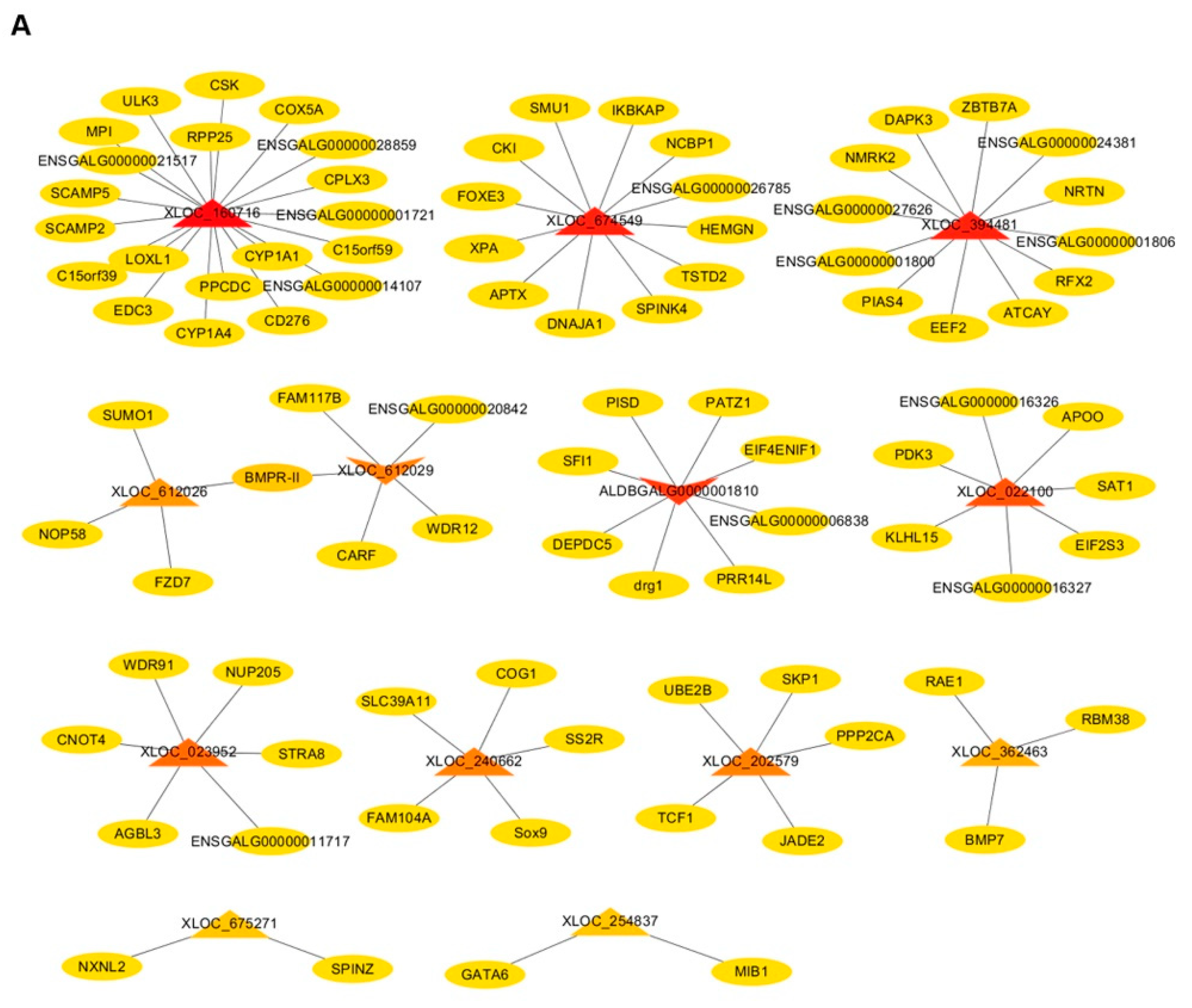
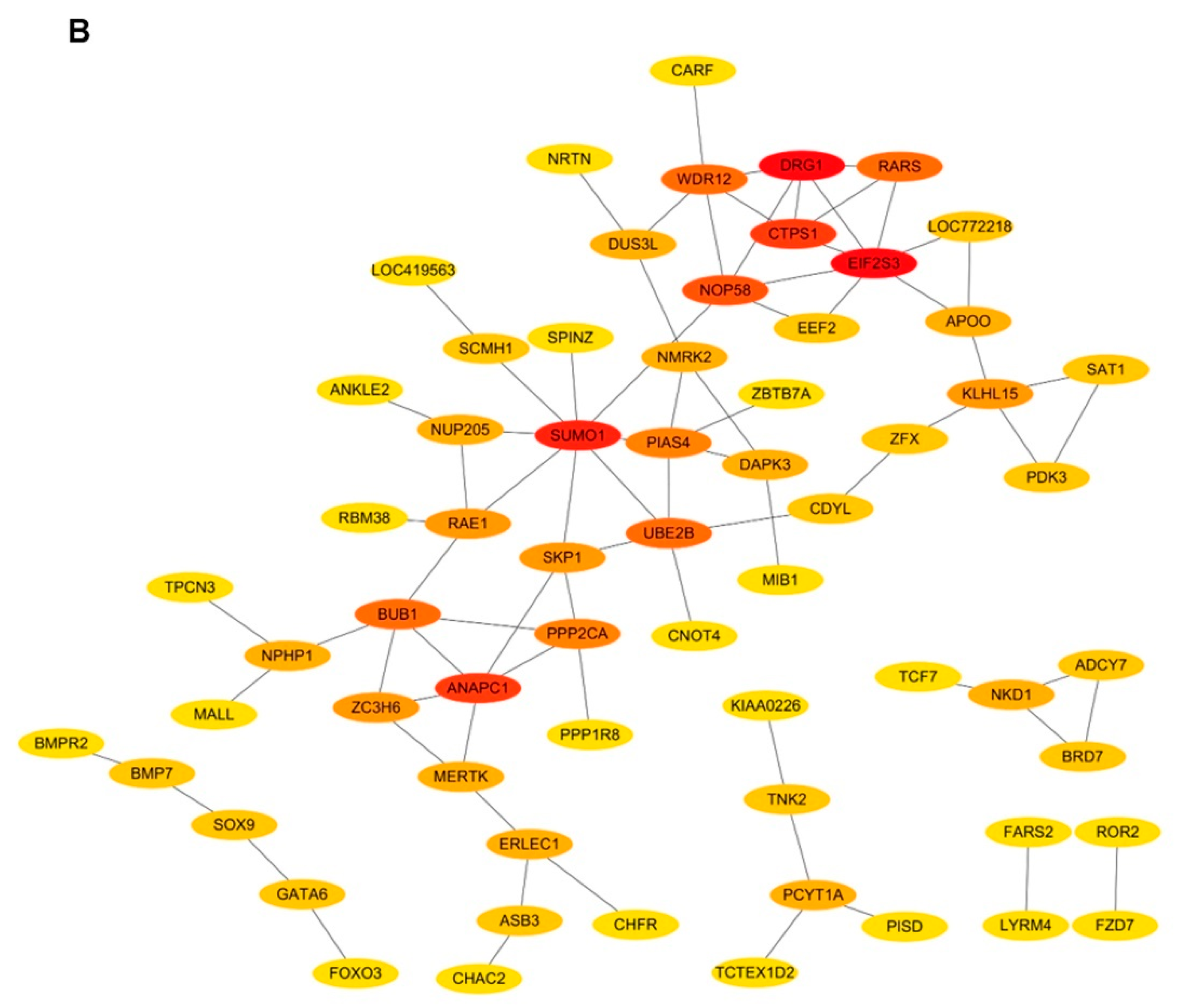
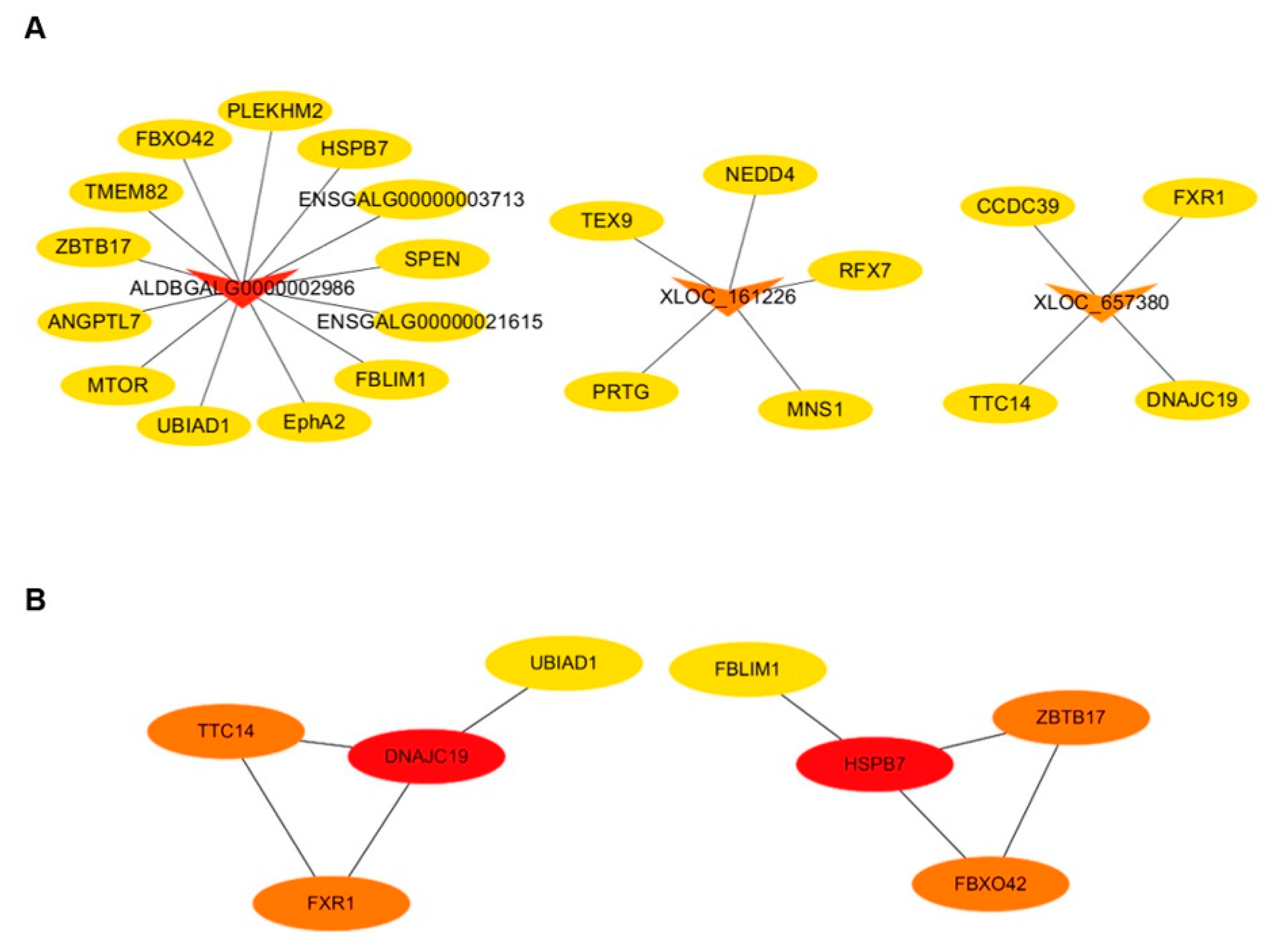
3.5. Co-expression Network Analysis of lncRNA–mRNA and mRNA–mRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Ramathal, C.; Pera, R.R.; Turek, P. Embryonic stem cells and the germ cell lineage. In Embryonic Stem Cells-Basic Biology to Bioengineering; Kallos, M., Ed.; InTech: London, UK, 2011; ISBN 978-953-307-278-4. Available online: http://www.intechopen.com/books/embryonic-stem-cells-basic-biology-tobioengineering/embryonic-stem-cells-and-the-germ-cell-lineage (accessed on 11 October 2020).
- Shah, S.M.; Saini, N.; Singh, M.K.; Manik, R.; Singla, S.K.; Palta, P.; Chauhan, M.S. Testicular cell–conditioned medium supports embryonic stem cell differentiation toward germ lineage and to spermatocyte- and oocyte-like cells. Theriogenology 2016, 86, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Lu, H.; Wu, L.T.; Liu, X.; Xiang, J.B. Retinoic acid can induce mouse embryonic stem cell R1/E to differentiate toward female germ cells while oleanolic acid can induce R1/E to differentiate toward both types of germ cells. Cell Biol. Int. 2014, 38, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-DiNardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Zhou, M.M.; Walsh, M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011, 71, 5365–5369. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Samira, B.; Wallace, H.A.; Russel, R.; Gholamreza, K. The therapeutic role of long non-coding RNAs in human diseases: A focus on the recent insights into autophagy. Pharmacol. Res. 2019, 142, 22–29. [Google Scholar]
- Chen, L.; Zhang, S. Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 2016, 366, 509–521. [Google Scholar] [CrossRef]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.H.; Wang, Y.M.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, L.; Xu, E.Y. LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development. Semin. Cell Dev. Biol. 2016, 59, 110–117. [Google Scholar] [CrossRef]
- Theresa, G.T.; Sargon, Y.; Jana, P.; Michal, R.F.; Jan, B.; Christian, R.; Christin, R.; Mehdi, G.; Martin, S.; Katsiaryna, T.; et al. The Vertebrate Protein Dead End Maintains Primordial Germ Cell Fate by Inhibiting Somatic Differentiation. Dev. Cell 2017, 43, 704–715. [Google Scholar]
- Zuo, Q.S.; Jin, J.; Jin, K.; Zhou, J.; Sun, C.; Song, J.Z.; Chen, G.G.; Zhang, Y.N.; Li, B.C. P53 and H3K4me2 activate N6-methylated LncPGCAT-1 to regulate primordial germ cell formation via MAPK signaling. J. Cell. Physiol. 2020, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, M.; Wang, M.; Wu, X.; Geng, L.; Xue, Y.; Wei, X.; Jia, Y.; Wu, X. A long non-coding RNA interacts with Gfra1 and maintains survival of mouse spermatogonial stem cells. Cell Death Dis. 2016, 7, e2140. [Google Scholar] [CrossRef] [PubMed]
- Weng, B.; Ran, M.L.; Chen, B.; He, C.Q.; Dong, L.H.; Peng, F.Z. Genome-wide analysis of long non-coding RNAs and their role in postnatal porcine testis development. Genomics 2017, 109, 446–456. [Google Scholar] [CrossRef]
- Hu, K.; Zhang, J.; Liang, M. LncRNA AK015322 promotes proliferation of spermatogonial stem cell C18-4 by acting as a decoy for microRNA-19b-3p. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 277–284. [Google Scholar] [CrossRef]
- Arun, G.; Akhade, V.S.; Donakonda, S.; Rao, M.R.S. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol. Cell. Biol. 2012, 32, 3140–3152. [Google Scholar] [CrossRef]
- Zuo, Q.S.; Zhang, C.; Jin, K.; Jing, J.; Sun, C.; Ahmed, M.F.; Song, J.Z.; Zhang, Y.N.; Chen, G.H.; Li, B.C. NICD-mediated notch transduction regulates the different fate of chicken primordial germ cells and spermatogonial stem cells. Cell Biosci. 2018, 8, 40. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A comparative genomic method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Matthew, D.Y.; Matthew, J.W.; Gordon, K.S.; Alicia, O. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta delta c(t) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, M.; Cheng, S.Z.; Zhang, C.; Wang, Y.L.; Zhang, W.H.; Zhao, R.F.; Sun, C.H.; Zhang, Y.N.; Li, B.C. CYP1A1 based on metabolism of xenobiotics by cytochrome P450 regulates chicken male germ cell differentiation. In Vitro Cell. Dev. Biol. Anim. 2017, 53, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Ferder, I.C.; Fung, L.; Ohguchi, Y.; Zhang, X.; Lassen, K.G.; Capen, D.; Brown, D.; Xavier, R.; Wang, N. Meiotic gatekeeper STRA8 suppresses autophagy by repressing Nr1d1 expression during spermatogenesis in mice. PLoS Genet. 2019, 15, e1008084. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.X.; Ma, L.F.; Lin, X.W.; Zhang, D.Q.; Li, Z.; Wu, Y.J.; Zheng, C.W.; Feng, X.; Liao, S.Y.; et al. Retinoic acid is sufficient for the in vitro induction of mouse spermatocytes. Stem Cell Rep. 2016, 7, 80–94. [Google Scholar] [CrossRef]
- Matsumoto, A.; Imagawa, E.; Miyake, N.; Ikeda, T.; Kobayashi, M.; Goto, M.; Matsumoto, N.; Yamagata, T.; Osaka, H. The presence of diminished white matter and corpus callosal thinning in a case with a SOX9 mutation. Brain Dev. 2018, 40, 325–329. [Google Scholar] [CrossRef]
- Ross, A.; Munger, S.; Capel, B. Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex. Dev. 2007, 1, 127–137. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Cheng, C.Y. Mammalian target of rapamycin complex (mTOR) pathway modulates blood-testis barrier (BTB) function through F-actin organization and gap junction. Histol. Histopathol. 2016, 31, 961. [Google Scholar] [PubMed]
- Xu, H.; Shen, L.J.; Chen, X.M.; Ding, Y.B.; He, J.L.; Zhu, J.; Wang, Y.X.; Liu, X.Q. mTOR/P70S6K promotes spermatogonia proliferation and spermatogenesis in Sprague Dawley rats. Reprod. Biomed. Online 2016, 32, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.S.; Li, T.P.; Ton, H.; Mao, X.D.; Chen, Y.J. Advances of Long Noncoding RNAs-mediated Regulation in Reproduction. Chin. Med. J. 2018, 131, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Tickle, C. The chicken as a model for embryonic development. Cytogenet. Genome Res. 2007, 117, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.Y.; Hao, Q.Y.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef]
- Saba, R.; Wu, Q.; Saga, Y. CYP26B1 promotes male germ cell differentiation by suppressing STRA8-dependent meiotic and STRA8-independent mitotic pathways. Dev. Biol. 2014, 389, 173–181. [Google Scholar] [CrossRef]
- Lee, H.C.; Lim, S.; Han, J.Y. Wnt/β-catenin signaling pathway activation is required for proliferation of chicken primordial germ cells in vitro. Sci. Rep. 2016, 6, 34510. [Google Scholar] [CrossRef]
- Lochab, A.K.; Extavour, C.G. Bone Morphogenetic Protein (BMP) signaling in animal reproductive system development and function. Dev. Biol. 2017, 427, 258–269. [Google Scholar] [CrossRef]
- Shen, C.; Yu, J.; Zhang, X.; Liu, C.C.; Guo, Y.S.; Zhu, J.W.; Zhang, K.; Yu, Y.; Gao, T.T.; Yang, S.M.; et al. Strawberry Notch 1 (SBN01) promotes proliferation of spermatogonial stem cells via the noncanonical Wnt pathway in mice. Asian J. Androl. 2019, 21, 345–350. [Google Scholar] [PubMed]
- Ni, F.D.; Hao, S.L.; Yang, W.X. Multiple signaling pathways in Sertoli cells: Recent findings in spermatogenesis. Cell Death Dis. 2019, 10, 541. [Google Scholar] [CrossRef]
- Stine, R.R.; Matunis, E.L. JAK-STAT signaling in stem cells. Adv. Exp. Med. Biol. 2013, 786, 247–267. [Google Scholar] [PubMed]
- Kee, K.; Reijo Pera, R.A. Human germ cell lineage differentiation from embryonic stem cells. CSH Protoc. 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, R.; Montanari, M.; Chiarella, P.; Stefanini, M.; Boitani, C. Regulatory role of BMP2 and BMP7 in spermatogonia and Sertoli cell proliferation in the immature mouse. Eur. J. Endocrinol. 2004, 151, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.F.; Parker, K.; Yao, H.H. WNT4/beta-catenin pathway maintains female germ cell survival by inhibiting activin betaB in the mouse fetal ovary. PLoS ONE 2010, 5, e10382. [Google Scholar] [CrossRef]
- He, N.; Wang, Y.; Zhang, C.; Wang, M.; Wang, Y.; Qysheng, Z.; Zhang, Y.; Bichun, L. Wnt signaling pathway regulates differentiation of chicken embryonic stem cells into spermatogonial stem cells via Wnt5a. J. Cell. Biochem. 2018, 119, 1689–1701. [Google Scholar] [CrossRef]
- Panneerdoss, S.; Viswanadhapalli, S.; Abdelfattah, N.; Onyeagucha, B.C.; Timilsina, S.; Mohammad, T.A.; Chen, Y.H.; Drake, M.; Vuori, K.; Kumar, T.R.; et al. Cross-talk between miR-471-5p and autophagy component proteins regulates LC3-associated phagocytosis (LAP) of apoptotic germ cells. Nat. Commun. 2017, 19, 598. [Google Scholar] [CrossRef]
- Wei, R.; Zhang, X.Y.; Cai, Y.H.; Liu, H.Y.; Wang, B.Y.; Zhao, X.D.; Zou, K. Busulfan Suppresses Autophagy in Mouse Spermatogonial Progenitor Cells via mTOR of AKT and p53 Signaling Pathways. Stem Cell Rev. Rep. 2020. [Google Scholar] [CrossRef]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- Cossec, J.C.; Theurillat, I.; Chica, C.; Aguín, S.B.; Gaume, X.; Andrieux, A.; Iturbide, A.; Jouvion, G.; Li, H.; Bossis, G.; et al. SUMO Safeguards Somatic and Pluripotent Cell Identities by Enforcing Distinct Chromatin States. Cell Stem Cell 2018, 23, 742–757. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Yun, S.M.; Jo, C.; Lee, D.H.; Choi, K.J.; Song, J.C.; Park, S.I.; Kim, Y.J.; Koh, Y.H. SUMO1 promotes Aβ production via the modulation of autophagy. Autophagy 2015, 11, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.J.; Guo, C.; Lao, Y.; Yang, J.; Chen, F.; Zhao, Y.Z.; Yang, Y.; Yang, J.; Yi, J. A fine-tuning mechanism underlying self-control for autophagy: deSUMOylation of BECN1 by SENP3. Autophagy 2020, 16, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Vigodner, M.; Ishikawa, T.; Schlegel, P.N.; Morris, P. SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am. J. Physiol. Endocrinol. Metab. 2006, 290, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Westman, B.J.; Verheggen, C.; Hutten, S.; Lam, Y.W.; Bertrand, E.; Lamond, A.I. A Proteomic Screen for Nucleolar SUMO Targets Shows SUMOylation Modulates the Function of Nop5/Nop58. Mol. Cell 2010, 39, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Pangas, S.A. Regulation of germ cell function by SUMOylation. Cell Tissue Res. 2016, 363, 47–55. [Google Scholar] [CrossRef]




| Long Non-Coding RNA/mRNA | Forward Primers (5′–3′) | Reverse Primers (5′–3′) | Product Size (bp) |
|---|---|---|---|
| ALDBGALT0000000220 | GTTAAGTGCAGCAAGACT | GAAGATGAGAACCAGAAA | 110 |
| TCONS_01079781 | AAGTACTAGAAGCCAGCT | GTTCTGTCTTAGCGATGT | 81 |
| TCONS_00005104 | AGAAGTCAGTGAAGAGCAGGAA | TCAAGCCCAGAACCCAAA | 134 |
| TCONS_00011769 | AAACGGATAATGACAAGA | ATTGCTTTCCCTGTATTT | 115 |
| TCONS_00277481 β-actin | AATAAATACTTCGGGTGA CAGCCATCTTTCTTGGGTAT | CTTTGACGTACTTTGTGC CTGTGATCTCCTTCTGCATCC | 150 169 |
| Sample Name | Raw Reads | Clean Reads | Clean Bases | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| ESC | 195781306 | 126200470 | 18.93G | 0.03 | 96.60 | 92.38 | 48.85 |
| PGC | 170137682 | 142487396 | 21.27G | 0.02 | 96.95 | 93.02 | 47.83 |
| SSC | 143227516 | 106333040 | 15.80G | 0.03 | 95.63 | 90.78 | 48.62 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, W.; Zhang, C.; Jin, K.; Zhang, Y.; Zuo, Q.; Li, B. Analysis of lncRNA Expression Profile during the Formation of Male Germ Cells in Chickens. Animals 2020, 10, 1850. https://doi.org/10.3390/ani10101850
Gao W, Zhang C, Jin K, Zhang Y, Zuo Q, Li B. Analysis of lncRNA Expression Profile during the Formation of Male Germ Cells in Chickens. Animals. 2020; 10(10):1850. https://doi.org/10.3390/ani10101850
Chicago/Turabian StyleGao, Wen, Chen Zhang, Kai Jin, Yani Zhang, Qisheng Zuo, and Bichun Li. 2020. "Analysis of lncRNA Expression Profile during the Formation of Male Germ Cells in Chickens" Animals 10, no. 10: 1850. https://doi.org/10.3390/ani10101850
APA StyleGao, W., Zhang, C., Jin, K., Zhang, Y., Zuo, Q., & Li, B. (2020). Analysis of lncRNA Expression Profile during the Formation of Male Germ Cells in Chickens. Animals, 10(10), 1850. https://doi.org/10.3390/ani10101850






