Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals, Farms and Sampling
2.3. Milk Analysis
2.4. DNA and Haplotype Analyses
2.5. Statistical Analyses
3. Results
3.1. Allele Frequencies at SPP1, POFUT1 and PRLR, and Association Analysis with Milk Traits
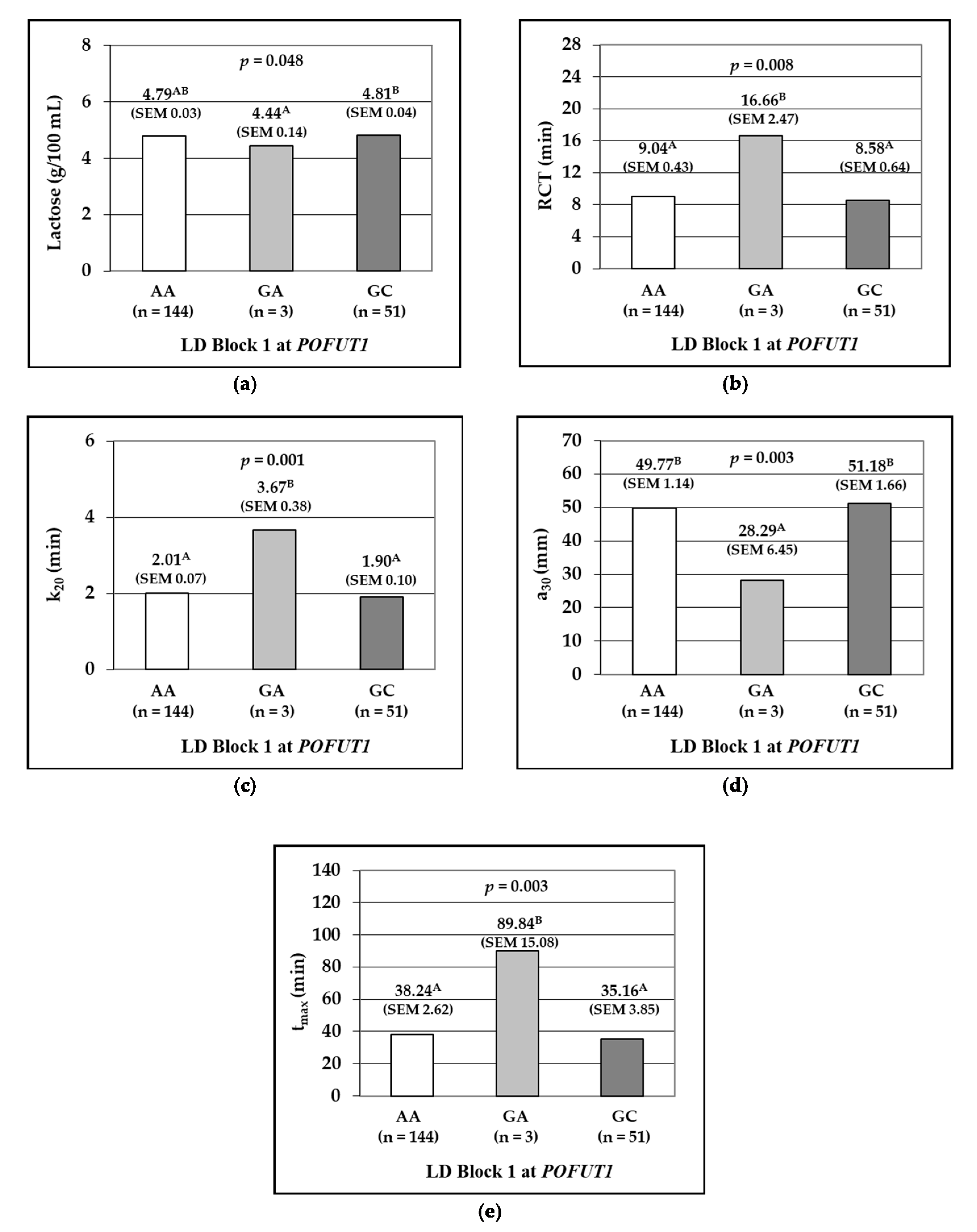
3.2. Linkage Disequilibrium and Association Analysis between Haplotype Blocks and Milk Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Statistical Database of the Food and Agriculture Organization of the United Nations. Available online: http://www.faostat.fao.org (accessed on 26 March 2020).
- Scintu, M.F.; Piredda, G. Typicity and biodiversity of goat and sheep milk products. Small Rumin. Res. 2007, 68, 221–231. [Google Scholar] [CrossRef]
- Balthazar, C.F.; Pimentel, T.C.; Ferrão, L.L.; Almada, C.N.; Santillo, A.; Albenzio, M.; Mollakhalili, N.; Mortazavian, A.M.; Nascimento, J.S.; Silva, M.C.; et al. Sheep milk: Physicochemical characteristics and relevance for functional food development. Compr. Rev. Food Sci. Food Saf. 2017, 16, 247–262. [Google Scholar] [CrossRef]
- Pazzola, M. Review: Coagulation traits of sheep and goat milk. Animals 2019, 9, 540. [Google Scholar] [CrossRef]
- Leroux, C.; Mazure, N.; Martin, P. Mutations away from splice site recognition sequences might cis-modulate alternative splicing of goat alpha sl-casein transcripts. J. Biol. Chem. 1992, 267, 6147–6157. [Google Scholar] [PubMed]
- Vacca, G.M.; Dettori, M.L.; Piras, G.; Manca, F.; Paschino, P.; Pazzola, M. Goat casein genotypes are associated with molk production traits in the Sarda breed. Anim. Genet. 2014, 45, 723–731. [Google Scholar] [CrossRef]
- Noce, A.; Pazzola, M.; Dettori, M.L.; Amills, M.; Castelló, A.; Cecchinato, A.; Bittante, G.; Vacca, G.M. Variations at regulatory regions of the milk protein genes are associated with milk traits and coagulation properties in the Sarda sheep. Anim. Genet. 2016, 47, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Dettori, M.L.; Pazzola, M.; Paschino, P.; Amills, M.; Vacca, G.M. Association between the GHR, GHRHR and IGF1 gene polymorphisms and milk yield and quality traits in Sarda sheep. J. Dairy Sci. 2018, 101, 9978–9986. [Google Scholar] [CrossRef]
- Dettori, M.L.; Pazzola, M.; Pira, E.; Stocco, G.; Vacca, G.M. Association between the GHR, GHRHR and IGF1 gene polymorphisms and milk coagulation properties in Sarda sheep. J. Dairy Res. 2019, 86, 331–336. [Google Scholar] [CrossRef]
- Kambadur, R.; Sharma, M.; Smith, T.P.L.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–915. [Google Scholar] [CrossRef]
- Gutièrrez-Gil, B.; Jose Arranz, J.; Pong-Wong, R.; García-Gámez, E.; Kijas, J.; Wiener, P. Application of selection mapping to identify genomic regions associated with dairy production in sheep. PLoS ONE 2014, 9, e94623. [Google Scholar] [CrossRef]
- Prince, C.W.; Oosawa, T.; Butler, W.T.; Tomana, M.; Bhown, A.S.; Bhown, M.; Schrohenloher, R. Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone. J. Biol. Chem. 1987, 262, 2900–2907. [Google Scholar]
- Erikson, D.W.; Burghardt, R.C.; Bayless, K.J.; Johnson G., A. Secreted phosphoprotein 1 (SPP1, Osteopontin) binds to integrin alphavbeta6 on porcine trophectoderm cells and integrin alphavbeta3 on uterine luminal epithelial cells, and promotes trophectoderm cell adhesion and migration. Biol. Reprod. 2009, 81, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.A.; Burghardt, R.C.; Bazer, F.W. Osteopontin: A leading candidate adhesion molecule for implantation in pigs and sheep. J. Anim. Sci. Biotechno. 2014, 5, 56. [Google Scholar] [CrossRef]
- Sheehy, P.A.; Riley, L.G.; Raadsma, H.W.; Williamson, P.; Wynn, P.C. A functional genomics approach to evaluate candidate genes located in a QTL interval for milk production traits on BTA6. Anim. Genet. 2009, 40, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Alain, K.; Karrow, N.A.; Thibault, C.; St-Pierre, J.; Lessard, M.; Bissonnette, N. Osteopontin: An early innate immune marker of Escherichia coli mastitis harbors genetic polymorphisms with possible links with resistance to mastitis. BMC Genom. 2009, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, N. Short communication: Genetic association of variations in the osteopontin gene (SPP1) with lactation persistency in dairy cattle. J. Dairy Sci. 2018, 101, 456–461. [Google Scholar] [CrossRef]
- Loriol, C.; Dupuy, F.; Rampal, R.; Dlugosz, M.A.; Haltiwanger, R.S.; Maftah, A.; Germot, A. Molecular evolution of protein O-fucosyltransferase genes and splice variants. Glycobiology 2006, 16, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P. Regulation of Notch signaling by glycosylation. Curr. Opin. Struc. Biol. 2007, 17, 530–535. [Google Scholar] [CrossRef]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Bole-Feysot, C.; Goffin, V.; Edery, M.; Binart, N.; Kelly, P.A. Prolactin (PRL) and its receptor: Actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr. Rev. 1998, 19, 225–268. [Google Scholar] [CrossRef]
- Ozmen, O.; Seker, I.; Ertugrul, O.; Ozkan, E.; Tekin, N. Prolactin receptor (PRLR) gene polymorphism in Chios, White Karaman and Awassi sheep breeds. Arch. Tierz. 2011, 54, 381–390. [Google Scholar]
- Wang, L.P.; Geng, R.Q.; Zhang, X.N.; Sun, W. Identification of SNPs within the PRLR gene and effects on maternal behavior in sheep. Genet. Mol. Res. 2015, 14, 17536–17543. [Google Scholar] [CrossRef]
- Szczesna, M.; Kirsz, K.; Misztal, T.; Zieba, D.A. Pregnancy-induced changes in the transcript levels of prolactin receptor and its suppressor in the ovine hypothalamus and adenohypophysis. Reprod. Dom. Anim. 2020, 55, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Pazzola, M.; Dettori, M.L.; Cipolat-Gotet, C.; Cecchinato, A.; Bittante, G.; Vacca, G.M. Phenotypic factors affecting coagulation properties of milk from Sarda ewes. J. Dairy Sci. 2014, 97, 7247–7257. [Google Scholar] [CrossRef] [PubMed]
- Vacca, G.M.; Pazzola, M.; Dettori, M.L.; Pira, E.; Malchiodi, F.; Cipolat-Gotet, C.; Cecchinato, A.; Bittante, G. Modeling of coagulation, curd firming, and syneresis of milk from Sarda ewes. J. Dairy Sci. 2015, 98, 2245–2259. [Google Scholar] [CrossRef]
- ISO-IDF (International Organization for Standardization and International Dairy Federation). Milk and Liquid Milk Products: Determination of Fat, Protein, Casein, Lactose and pH Content; International Standard ISO 9622 and IDF, 141:2013; ISO: Geneva, Switzerland; IDF: Brussels, Belgium, 2013. [Google Scholar]
- ISO-IDF (International Organization for Standardization and International Dairy Federation). Milk: Quantitative Determination of Bacteriological Quality—Guidance for Establishing and Verifying a Conversion Relationship between Routine Method Results and Anchor Method Results; International Standard ISO 21187 and IDF, 196:2004; ISO: Geneva, Switzerland; IDF: Brussels, Belgium, 2004. [Google Scholar]
- ISO-IDF (International Organization for Standardization and International Dairy Federation). Milk: Bacterial Count—Protocol for the Evaluation of Alternative Methods; International Standard ISO 16297:2013 and IDF, 161:2013; ISO: Geneva, Switzerland; IDF: Brussels, Belgium, 2013. [Google Scholar]
- ISO-IDF (International Organization for Standardization and International Dairy Federation). Milk: Enumeration of Somatic Cells—Part 2: Guidance on the Operation of Fluoro-Opto-Electronic Counters; International Standard ISO 13366-2 and IDF IDF, 148-2:2006; ISO: Geneva, Switzerland; IDF: Brussels, Belgium, 2006. [Google Scholar]
- Shook, G.E. Genetic improvement of mastitis through selection on somatic cell count. Vet. Clin. N. Am. Food Anim. Pract. 1993, 9, 563–577. [Google Scholar] [CrossRef]
- Mc Mahon, D.J.; Brown, R.J. Evaluation of Formagraph for comparing rennet solutions. J. Dairy Sci. 1982, 65, 1639–1642. [Google Scholar] [CrossRef]
- Bittante, G. Modeling rennet coagulation and curd firmness of milk. J. Dairy Sci. 2011, 94, 5821–5832. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef]
- Dudemaine, P.L.; Thibault, C.; Alain, K.; Bissonnette, N. Genetic variations in the SPP1 promoter affect gene expression and the level of osteopontin secretion into bovine milk. Anim. Genet. 2014, 45, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Larrañaga, O.; Langa, J.; Rendo, F.; Manzano, C.; Iriondo, M.; Estonba, A. Genomic selection signatures in sheep from the Western Pyrenees. Genet. Sel. Evol. 2018, 50, 9. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, A.A.; Deniskova, T.E.; Yudin, N.S.; Dotsev, A.V.; Khamiruev, T.N.; Selionova, M.I.; Egorov, S.V.; Reyer, H.; Wimmers, K.; Brem, G.; et al. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genom. 2019, 20, 294. [Google Scholar] [CrossRef] [PubMed]
- Kułaj, D.; Pokorska, J.; Ochrem, A.; Dusza, M.; Makulska, J. Effects of the c.8514C > T polymorphism in the osteopontin gene (OPN) on milk production, milk composition and disease susceptibility in Holstein-Friesian cattle. It. J. Ani. Sci. 2019, 18, 546–553. [Google Scholar] [CrossRef]
- Mello, F.; Cobuci, J.A.; Martins, M.F.; Silva, M.V.G.B.; Neto, J.B. Association of the polymorphism g. 8514C>T in the osteopontin gene (SPP1) with milk yield in the dairy cattle breed Girolando. Anim. Genet. 2012, 43, 647–648. [Google Scholar] [CrossRef]
- Garcia-Fernandez, M.; Gutierrez-Gil, B.; Sanchez, J.P.; Moran, J.A.; Garcia-Gamez, E.; Alvarez, L.; Arranz, J.J. The role of bovine causal genes underlying dairy traits in Spanish Churra sheep. Anim. Genet. 2011, 42, 415–420. [Google Scholar] [CrossRef]
- Paschino, P.; Vacca, G.M.; Dettori, M.L.; Pazzola, M. An approach for the estimation of somatic cells’ effect in Sarda sheep milk based on the analysis of milk traits and coagulation properties. Small Rumin. Res. 2019, 171, 77–81. [Google Scholar] [CrossRef]
- Pazzola, M.; Cipolat-Gotet, C.; Bittante, G.; Cecchinato, A.; Dettori, M.L.; Vacca, G.M. Phenotypic and genetic relationships between indicators of the mammary gland health status and milk composition, coagulation, and curd firming in dairy sheep. J. Dairy Sci. 2018, 101, 3164–3175. [Google Scholar] [CrossRef]
- Lucey, J.A. Rennet-induced coagulation of milk. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Fox, P.F., McSweeney, P.L.H., Eds.; Academic Press: San Diego, CA, USA, 2011; Volume 1, pp. 579–584. [Google Scholar]
- Ogorevc, J.; Kunej, T.; Razpet, A.; Dovc, P. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim. Genet. 2009, 40, 832–851. [Google Scholar] [CrossRef]
- Buono, K.D.; Robinson, G.W.; Martin, C.; Shi, S.; Stanley, P.; Tanigaki, K.; Honjo, T.; Hennighausen, L. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev. Biol. 2006, 293, 565–580. [Google Scholar] [CrossRef]
- Bittante, G.; Contiero, B.; Cecchinato, A. Prolonged observation and modelling of milk coagulation, curd firming, and syneresis. Int. Dairy J. 2013, 29, 115–123. [Google Scholar] [CrossRef]
- Dadousis, C.; Biffani, S.; Cipolat-Gotet, C.; Nicolazzi, E.L.; Rossoni, A.; Santus, E.; Bittante, G.; Cecchinato, A. Genome-wide association of coagulation properties, curd firmness modeling, protein percentage, and acidity in milk from Brown Swiss cows. J. Dairy Sci. 2016, 99, 3654–3666. [Google Scholar] [CrossRef] [PubMed]
- Dadousis, C.; Pegolo, S.; Rosa, G.J.M.; Gianola, D.; Bittante, G.; Cecchinato, A. Pathway-based genome-wide association analysis of milk coagulation properties, curd firmness, cheese yield, and curd nutrient recovery in dairy cattle. J. Dairy Sci. 2017, 100, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Cassy, S.; Charlier, M.; Belair, L.; Guillomot, M.; Charron, G.; Bloch, B.; Djiane, J. Developmental expression and localization of the prolactin receptor (PRL-R) gene in ewe mammary gland during pregnancy and lactation: Estimation of the ratio of the two forms of PRL-R messenger ribonucleic acid. Biol. Reprod. 1998, 58, 1290–1296. [Google Scholar] [CrossRef] [PubMed]


| Genes and SNP ID | Chr pos. | Gene Region | ObsH | PredH | HWpv | %Gen | MAF | Alleles |
|---|---|---|---|---|---|---|---|---|
| SPP1 | ||||||||
| rs161844011 | OAR6:36651870 | exon 7 1 | 0.434 | 0.468 | 0.202 | 98.1 | 0.373 | (T):C |
| rs426249393 | OAR6:36658163 | exon 1 2 | 0.384 | 0.394 | 0.672 | 99.2 | 0.27 | A:(G) |
| POFUT1 | ||||||||
| rs424501869 | OAR13:61007495 | intron 1 | 0.446 | 0.476 | 0.266 | 100.0 | 0.39 | A:(G) |
| rs421284407 | OAR13:61009391 | intron 2 | 0.0 | 0.0 | 1.0 | 93.3 | 0.0 | C |
| rs408068827 | OAR13:61013709 | intron 3 | 0.422 | 0.454 | 0.206 | 100.0 | 0.348 | A:(C) |
| PRLR | ||||||||
| rs412695065 | OAR16:38969344 | intron 1 | 0.402 | 0.370 | 0.334 | 100.0 | 0.245 | A:(C) |
| rs400874750 | OAR16:39004070 | intron 2 | 0.538 | 0.488 | 0.223 | 100.0 | 0.421 | T:(C) |
| rs428472303 | OAR16:39006813 | intron 2 | 0.506 | 0.467 | 0.3585 | 96.7 | 0.371 | T:(C) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dettori, M.L.; Pazzola, M.; Petretto, E.; Vacca, G.M. Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep. Animals 2020, 10, 1216. https://doi.org/10.3390/ani10071216
Dettori ML, Pazzola M, Petretto E, Vacca GM. Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep. Animals. 2020; 10(7):1216. https://doi.org/10.3390/ani10071216
Chicago/Turabian StyleDettori, Maria Luisa, Michele Pazzola, Elena Petretto, and Giuseppe Massimo Vacca. 2020. "Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep" Animals 10, no. 7: 1216. https://doi.org/10.3390/ani10071216
APA StyleDettori, M. L., Pazzola, M., Petretto, E., & Vacca, G. M. (2020). Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep. Animals, 10(7), 1216. https://doi.org/10.3390/ani10071216





