Ancient Homozygosity Segments in West African Djallonké Sheep Inform on the Genomic Impact of Livestock Adaptation to the Environment
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Available
2.2. Population Structure Analyses
2.3. Detection of Runs of Homozygosity
2.4. Homozygosity-by-Descent Analyses
2.5. Candidate Homozygous Segments and Enrichment and Functional Annotation Analyses
3. Results
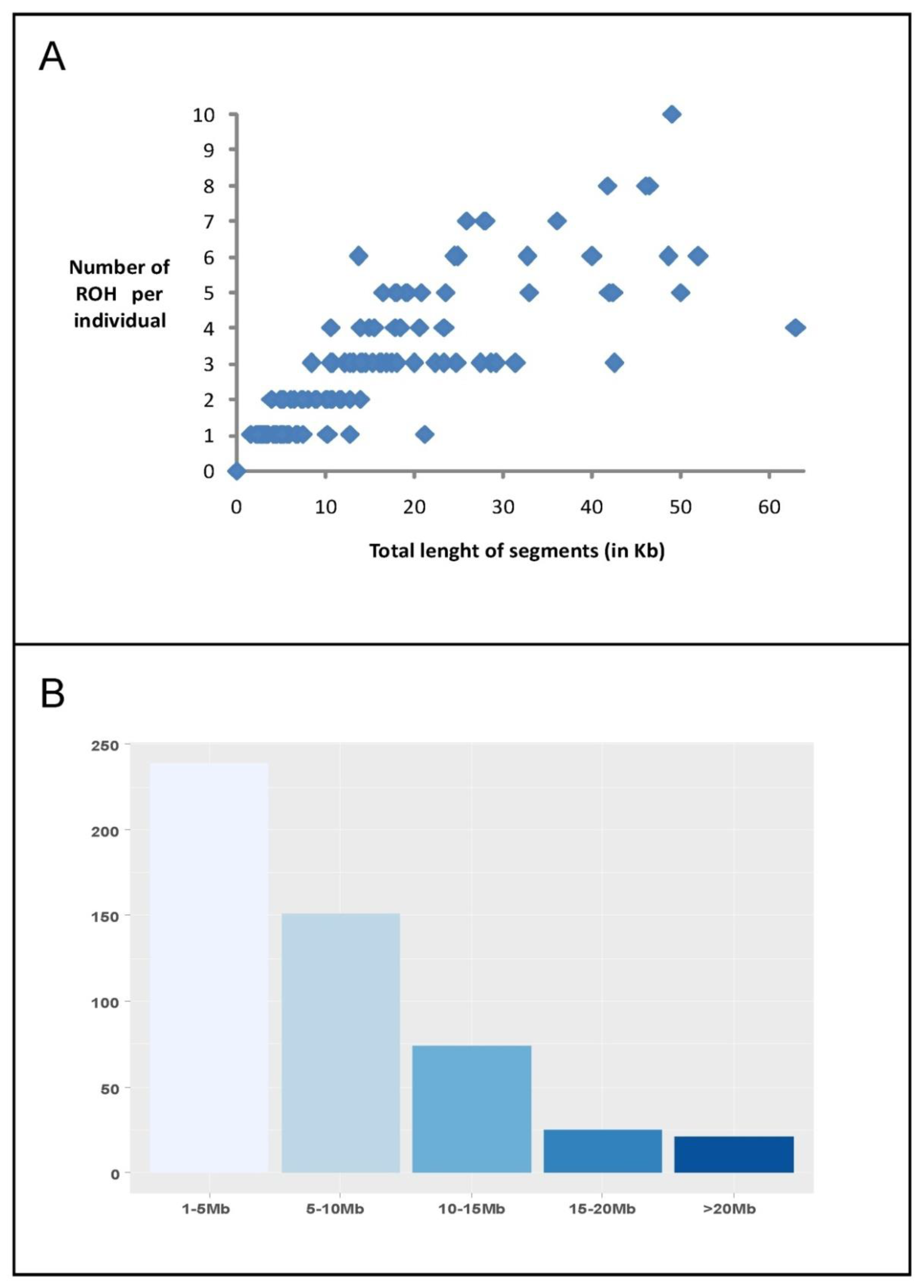
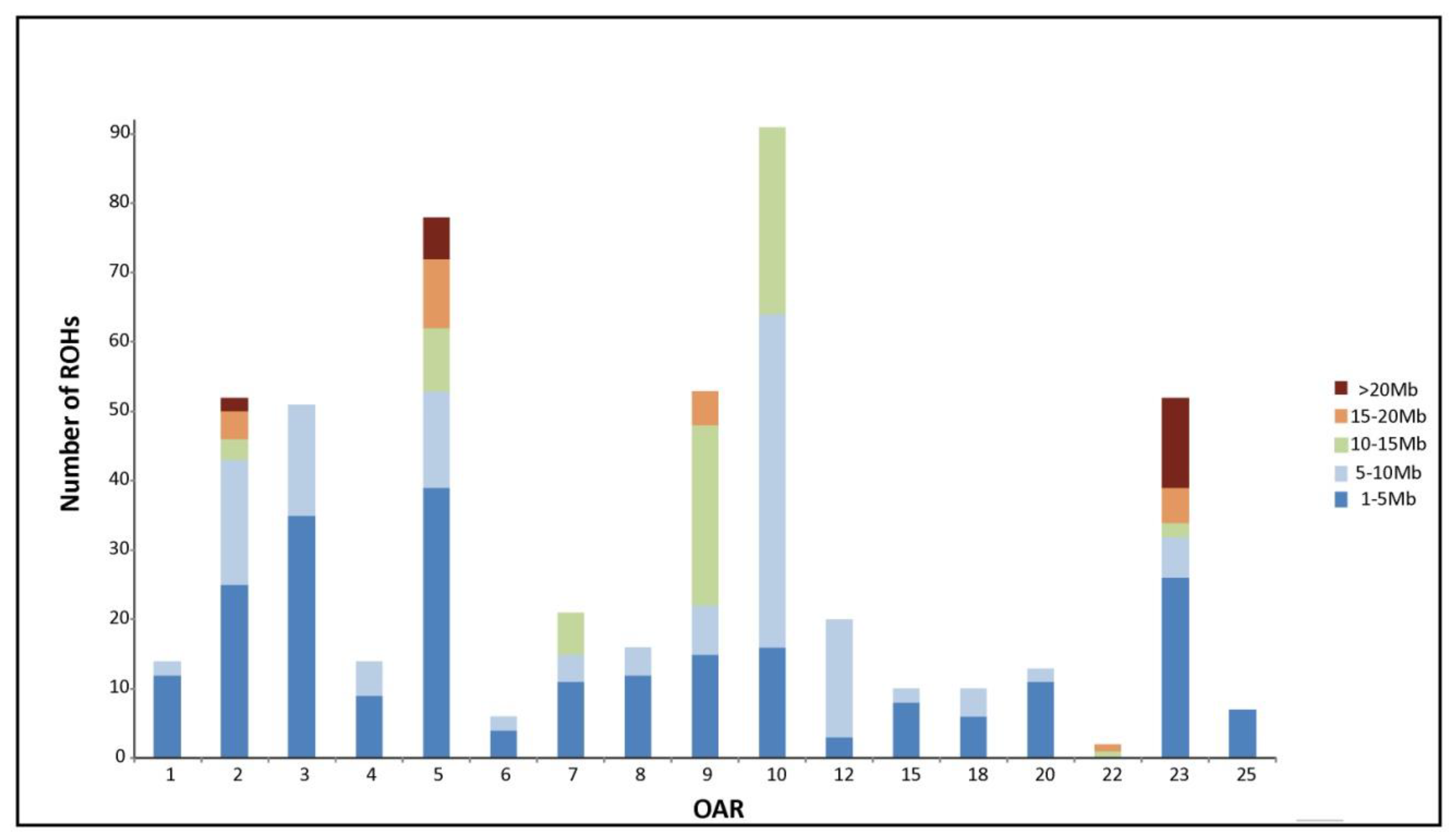
3.1. Genomic Homozygosity Distribution
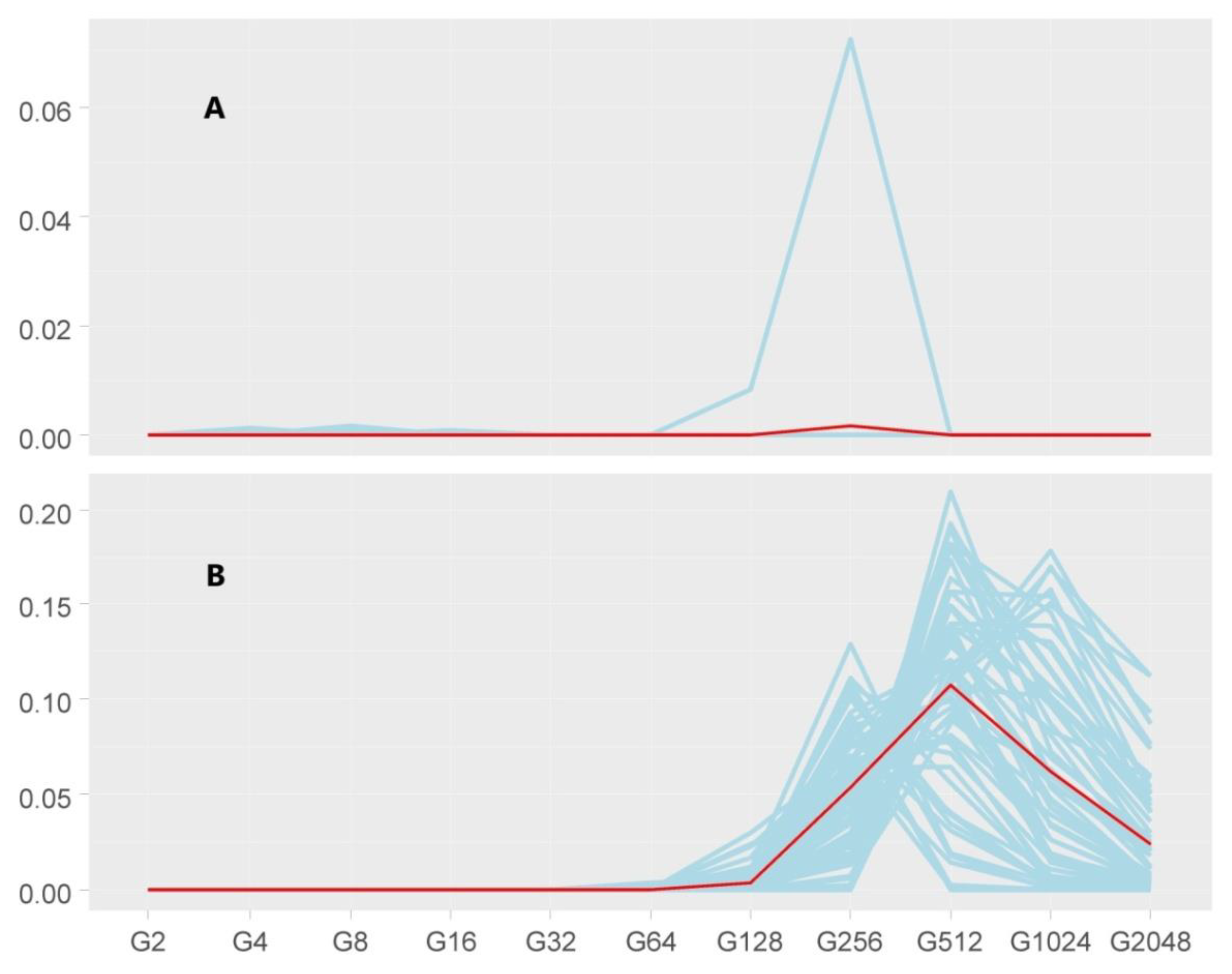
3.2. Homozygosity-by-Descent Analyses
3.3. ROH-HBD Intersections and Identification of Functional Candidate Genes
4. Discussion
4.1. Homozygosity and Autozygosity in Djallonké Sheep
4.2. Biological Importance of the Functional Clusters Identified
4.3. Consistency with Previous Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.; et al. Revealing the history of sheep domestication using retrovirus integrations. Science 2009, 324, 532–536. [Google Scholar] [CrossRef]
- Muigai, A.W.T.; Hanotte, O. The origin of African sheep: Archaeological and genetic perspectives. Afr. Archaeol. Rev. 2013, 30, 39–50. [Google Scholar] [CrossRef]
- Gifford-Gonzalez, D. Animal Disease Challenges to the Emergence of Pastoralism in Sub-Saharan Africa. Afr. Archaeol. Rev. 2000, 17, 95–139. [Google Scholar] [CrossRef]
- Traoré, A.; Tamboura, H.H.; Kaboré, A.; Royo, L.J.; Fernández, I.; Álvarez, I.; Sangare, M.; Bouchel, D.; Poivey, J.P.; Francois, D.; et al. Multivariate characterization of morphological traits in Burkina Faso sheep. Small Rum. Res. 2008, 80, 62–67. [Google Scholar] [CrossRef]
- Álvarez, I.; Traoré, A.; Tambourá, H.H.; Kaboré, A.; Royo, L.J.; Fernández, I.; Ouédraogo-Sanou, G.; Sawadogo, L.; Goyache, F. Microsatellite analysis characterizes Burkina Faso as a genetic contact zone between Sahelian and Djallonké sheep. Anim. Biotech. 2009, 20, 47–57. [Google Scholar] [CrossRef]
- Geerts, S.; Osaer, S.; Goossens, B.; Faye, D. Trypanotolerance in small ruminants of sub-Saharan Africa. Trends Parasitol. 2009, 25, 132–138. [Google Scholar] [CrossRef]
- Álvarez, I.; Fernández, I.; Traoré, A.; Pérez-Pardal, L.; Menéndez-Arias, N.A.; Goyache, F. Genomic scan of selective sweeps in Djallonké (West African Dwarf) sheep shed light on adaptation to harsh environments. Sci. Rep. 2020, 10, 2824. [Google Scholar] [CrossRef] [PubMed]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Bertolini, F.; Cardoso, T.F.; Marras, G.; Nicolazzi, E.L.; Rothschild, M.F.; Amills, M.; The AdaptMap Consortium. Genome-wide patterns of homozygosity provide clues about the population history and adaptation of goats. Genet. Sel. Evol. 2018, 50, 59. [Google Scholar] [CrossRef] [PubMed]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-Wide Characterization of Selection Signatures and Runs of Homozygosity in Ugandan Goat Breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.-J.; Madsen, O.; Paudel, Y.; Frantz, L.A.F.; Schook, L.B.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Purfield, D.C.; McParland, S.; Wall, E.; Berry, D.P. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Ferenčaković, M.; Hamzić, E.; Gredler, B.; Solberg, T.R.; Klemetsdal, G.; Curik, I.; Sölkner, J. Estimates of autozygosity derived from runs of homozygosity: Empirical evidence from selected cattle populations. J. Anim. Breed. Genet. 2013, 130, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Peripolli, E.; Stafuzza, N.B.; Amorim, S.T.; de Lemos, M.V.; Grigoletto, L.; Kluska, S.; Ferraz, J.B.S.; Pereira, J.; Chicaroni, E.; Mattos, E.; et al. Genome-wide scan for runs of homozygosity in the composite Montana Tropical beef cattle. J. Anim. Breed. Genet. 2020, 137, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Ferenčaković, M.; Sölkner, J.; Kaps, M.; Curik, I. Genome-wide mapping and estimation of inbreeding depression of semen quality traits in a cattle population. J. Dairy Sci. 2017, 100, 4721–4730. [Google Scholar] [CrossRef]
- Druet, T.; Gautier, M.A. Model-based approach to characterize individual inbreeding at both global and local genomic scales. Mol. Ecol. 2017, 26, 5820–5841. [Google Scholar] [CrossRef]
- Bertrand, A.R.; Kadri, N.K.; Flori, L.; Gautier, M.; Druet, T. RZooRoH: An R package to characterize individual genomic autozygosity and identify homozygous-by-descent segments. Methods Ecol. Evol. 2019, 10, 860–866. [Google Scholar] [CrossRef]
- Álvarez, I.; Traoré, A.; Fernández, I.; Soudré, A.; Kaboré, A.; Menéndez-Arias, N.A.; Sanou, M.; Tamboura, H.H.; Goyache, F. Usefulness of running animal models in absence of pedigrees: Estimation of genetic parameters for gastrointestinal parasite resistance traits in Djallonké sheep of Burkina Faso. Small Rum. Res. 2018, 161, 81–88. [Google Scholar] [CrossRef]
- Álvarez, I.; Fernández, I.; Soudré, A.; Traoré, A.; Pérez-Pardal, L.; Sanou, M.; Tapsoba, S.A.R.; Menéndez-Arias, N.A.; Goyache, F. Identification of genomic regions and candidate genes of functional importance for gastrointestinal parasite resistance traits in Djallonké sheep of Burkina Faso. Arch. Anim. Breed. 2019, 62, 313–323. [Google Scholar] [CrossRef]
- Traoré, A.; Notter, D.R.; Soudré, A.; Kaboré, A.; Álvarez, I.; Fernández, I.; Sanou, M.; Samshuddin, M.; Periassamy, K.; Tamboura, H.H.; et al. Resistance to gastrointestinal parasite infection in Djallonké sheep. Animal 2017, 11, 1354–1362. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning, A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; p. 1546. [Google Scholar]
- Traoré, A.; Álvarez, I.; Fernández, I.; Pérez-Pardal, L.; Kaboré, A.; Ouédraogo-Sanou, G.M.S.; Zaré, Y.; Tamboura, H.H.; Goyache, F. Ascertaining gene flow patterns in livestock populations of developing countries: A case study in Burkina Faso goat. BMC Genetics 2012, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4. [Google Scholar] [CrossRef]
- Manunza, A.; Noce, A.; Serradilla, J.M.; Goyache, F.; Martínez, A.; Capote, J.; Delgado, J.V.; Jordana, J.; Muñoz, E.; Molina, A.; et al. A genome-wide perspective about the diversity and demographic history of seven Spanish goat breeds. Genet. Sel. Evol. 2016, 48, 52. [Google Scholar] [CrossRef] [PubMed]
- Nandolo, W.; Mészáros, G.; Banda, L.J.; Gondwe, T.N.; Lamuno, D.; Mulindwa, H.A.; Nakimbugwe, H.N.; Wurzinger, M.; Utsunomiya, Y.T.; Woodward-Greene, M.J.; et al. Timing and Extent of Inbreeding in African Goats. Front. Genet. 2019, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Goyache, F.; Gutiérrez, J.P.; Fernández, I.; Gómez, E.; Álvarez, I.; Díez, J.; Royo, L.J. Using pedigree information to monitor genetic variability of endangered populations: The Xalda sheep breed of Asturias as an example. J. Anim. Breed. Genet. 2003, 120, 95–103. [Google Scholar] [CrossRef]
- Solé, M.; Gori, A.; Faux, P.; Bertrand, A.; Farnir, F.; Gautier, M.; Druet, T. Age-based partitioning of individual genomic inbreeding levels in Belgian Blue cattle. Genet. Sel. Evol. 2017, 49, 92. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. EnsemblBioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Yu, Y.; Ouyang, Y.; Yao, W. shinyCircos: An R/Shiny application for interactive creation of Circos plot. Bioinformatics 2018, 34, 1229–1231. [Google Scholar] [CrossRef]
- Ferenčaković, M.; Sölkner, J.; Curik, I. Estimating autozygosity from high-throughput information: Effects of SNP density and genotyping errors. Genet. Sel. Evol. 2013, 45, 42. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, F.C.; Joshi, P.K.; Clark, D.W.; Ramsay, M.; Wilson, J.F. Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 2018, 19, 220–234. [Google Scholar] [CrossRef]
- Ceballos, F.C.; Hazelhurst, S.; Ramsay, M. Runs of homozygosity in sub-Saharan African populations provide insights into complex demographic histories. Hum. Genet. 2019, 138, 1123–1142. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, T.J.; Absher, D.; Feldman, M.W.; Myers, R.M.; Rosenberg, N.A.; Li, J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012, 91, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Nothnagel, M.; Lu, T.; Kayser, M.; Krawczak, M. Genomic and geographic distribution of SNP-defined runs of homozygosity in Europeans. Hum. Mol. Genet. 2010, 19, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef]
- Irwin, D.M. Genomic organization and evolution of ruminant lysozyme c genes. Zool. Res. 2015, 36, 1–17. [Google Scholar] [CrossRef]
- Zani, I.A.; Stephen, S.L.; Mughal, N.A.; Russell, D.; Homer-Vanniasinkam, S.; Wheatcroft, S.B.; Ponnambalam, S. Scavenger receptor structure and function in health and disease. Cells 2015, 4, 178–201. [Google Scholar] [CrossRef]
- Guo, Z.; González, J.; Hernandez, J.; McNeilly, T.N.; Corripio-Miyar, Y.; Frew, D.; Morrison, T.; Yu, P.; Li, R.W. Possible mechanisms of host resistance to Haemonchus infection in sheep breeds native to the Canary Islands. Sci. Rep. 2016, 6, 26200. [Google Scholar] [CrossRef]
- Waluk, D.P.; Sucharski, F.; Sipos, L.; Silberring, J.; Hunt, M.C. Reversible lysine acetylation regulates activity of human glycine N-acyltransferase-like 2 (hGLYATL2): Implications for production of glycine-conjugated signaling molecules. J. Biol. Chem. 2012, 287, 16158–16167. [Google Scholar] [CrossRef]
- Schmidt, A.; Hall, A. Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes Dev. 2002, 16, 1587–1609. [Google Scholar] [CrossRef] [PubMed]

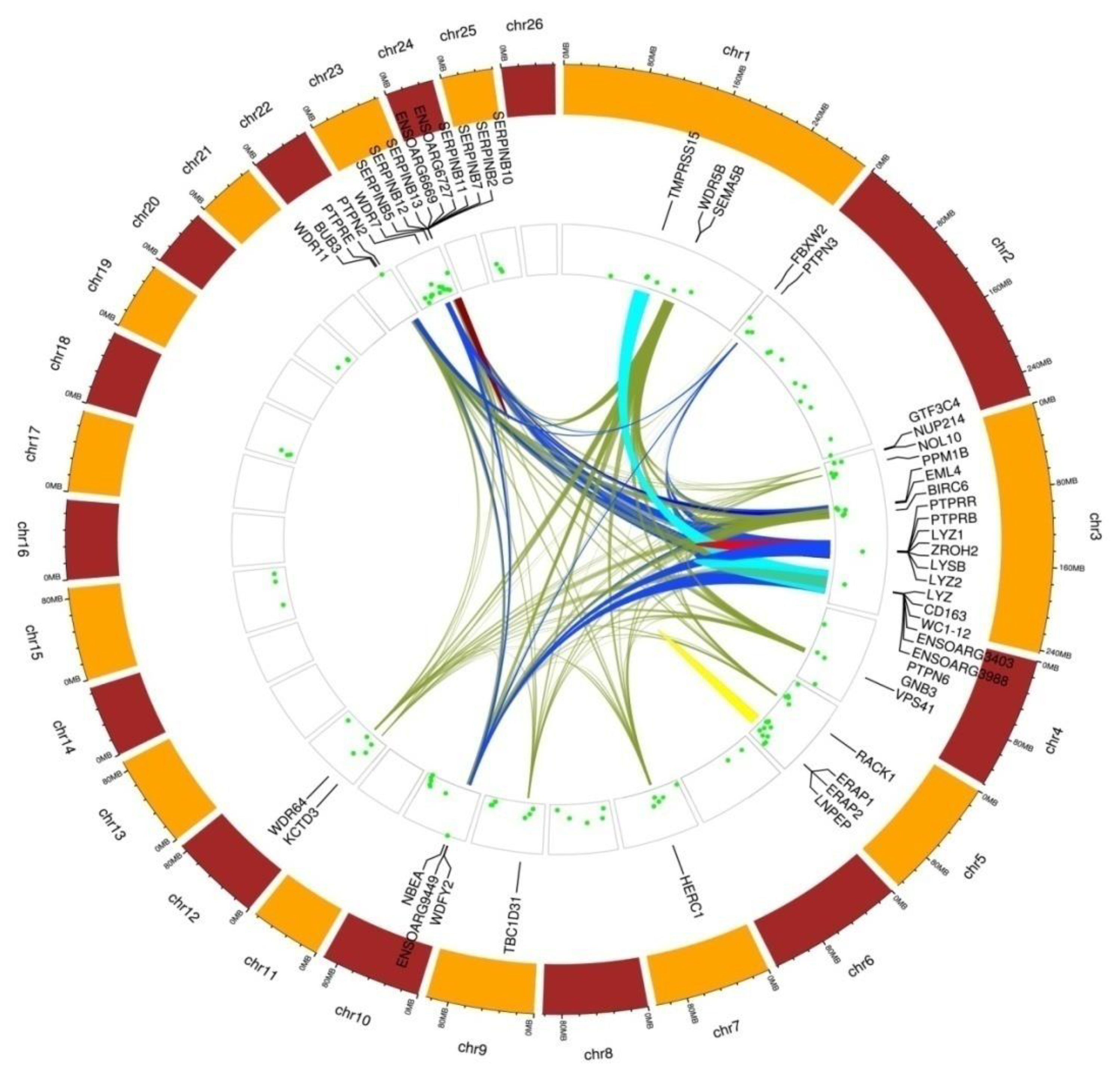
| Functional Cluster (Enrichment Score) | Term Description | Count | p-Value | |
|---|---|---|---|---|
| Cluster1 (3.084) | INTERPRO | IPR000215:Serpin family | 11 | 7.49 × 10−4 |
| INTERPRO | IPR023796:Serpin domain | 11 | 7.49 × 10−4 | |
| SMART | SM00093:SERPIN | 11 | 8.29 × 10−4 | |
| INTERPRO | IPR023795:Protease inhibitor I4, serpin, conserved site | 10 | 9.96 × 10−4 | |
| Cluster2 (2.024) | INTERPRO | IPR019799:Glycoside hydrolase, family 22, conserved site | 5 | 0.005281 |
| INTERPRO | IPR000974:Glycoside hydrolase, family 22, lysozyme | 5 | 0.007487 | |
| INTERPRO | IPR001916:Glycoside hydrolase, family 22 | 5 | 0.010224 | |
| SMART | SM00263:LYZ1 | 5 | 0.010761 | |
| GOTERM_MF_DIRECT | GO:0003796~lysozyme activity | 5 | 0.012172 | |
| INTERPRO | IPR023346:Lysozyme-like domain | 5 | 0.013533 | |
| Cluster3 (2.011) | INTERPRO | IPR001190:Speract/scavenger receptor | 8 | 0.004788 |
| SMART | SM00202:SR | 8 | 0.005171 | |
| INTERPRO | IPR017448:Speract/scavenger receptor-related | 8 | 0.008159 | |
| GOTERM_MF_DIRECT | GO:0005044~scavenger receptor activity | 8 | 0.044628 | |
| Cluster4 (2.008) | INTERPRO | IPR009122:Desmosomal cadherin | 7 | 7.62 × 10−7 |
| INTERPRO | IPR027397:Catenin binding domain | 7 | 0.005220 | |
| INTERPRO | IPR000233:Cadherin, cytoplasmic domain | 6 | 0.019394 | |
| INTERPRO | IPR002126:Cadherin | 10 | 0.036299 | |
| INTERPRO | IPR015919:Cadherin-like | 10 | 0.039529 | |
| SMART | SM00112:CA | 9 | 0.070844 | |
| GOTERM_BP_DIRECT | GO:0007156~homophilic cell adhesion via plasma membrane adhesion molecules | 10 | 0.094881 | |
| INTERPRO | IPR020894:Cadherin conserved site | 8 | 0.116413 | |
| Cluster5 (1.563) | INTERPRO | IPR010313:Glycine N-acyltransferase | 3 | 0.026686 |
| INTERPRO | IPR015938:Glycine N-acyltransferase, N-terminal | 3 | 0.02668622 | |
| INTERPRO | IPR013652:Glycine N-acyltransferase, C-terminal | 3 | 0.026686 | |
| GOTERM_MF_DIRECT | GO:0047961~glycine N-acyltransferase activity | 3 | 0.029378 | |
| Cluster6 (1.419) | INTERPRO | IPR000651:Ras-like guanine nucleotide exchange factor, N-terminal | 6 | 0.016082 |
| SMART | SM00229:RasGEFN | 5 | 0.028570 | |
| SMART | SM00147:RasGEF | 6 | 0.051153 | |
| INTERPRO | IPR001895:Guanine-nucleotide dissociation stimulator CDC25 | 6 | 0.054995 | |
| INTERPRO | IPR023578:Ras guanine nucleotide exchange factor, domain | 6 | 0.061954 | |
| Functional Cluster (Enrichment Score) | Gene Name | Description | EnsemblID | OAR | Gene Start | Gene End |
|---|---|---|---|---|---|---|
| Cluster1 (3.084) | SERPINB5 | serpin family B member 5 [Source:HGNC Symbol;Acc:HGNC:8949] | ENSOARG00000006391 | 23 | 61974470 | 61989454 |
| SERPINB12 | serpin family B member 12 [Source:HGNC Symbol;Acc:HGNC:14220] | ENSOARG00000006441 | 23 | 62028783 | 62041541 | |
| SERPINB13 | serpin family B member 13 [Source:HGNC Symbol;Acc:HGNC:8944] | ENSOARG00000006513 | 23 | 62045722 | 62057218 | |
| ENSOARG00000006669 | ENSOARG00000006669 | 23 | 62100351 | 62105829 | ||
| ENSOARG00000006727 | serpin B4-like [Source:NCBI gene;Acc:101104114] | ENSOARG00000006727 | 23 | 62117011 | 62125799 | |
| SERPINB11 | serpin family B member 11 (gene/pseudogene) [Source:HGNC Symbol;Acc:HGNC:14221] | ENSOARG00000006806 | 23 | 62162337 | 62177361 | |
| SERPINB7 | serpin family B member 7 [Source:HGNC Symbol;Acc:HGNC:13902] | ENSOARG00000006880 | 23 | 62189501 | 62220423 | |
| SERPINB2 | plasminogen activator inhibitor 2 [Source:NCBI gene;Acc:101105044] | ENSOARG00000006889 | 23 | 62287023 | 62298724 | |
| SERPINB10 | serpin family B member 10 [Source:HGNC Symbol;Acc:HGNC:8942] | ENSOARG00000006971 | 23 | 62308486 | 62330628 | |
| Cluster2 (2.024) | LYZ1 | Ovisarieslysozyme C-1-like (LOC443320), mRNA. [Source:RefSeq mRNA;Acc:NM_001308588] | ENSOARG00000020393 | 3 | 150160411 | 150294070 |
| ENSOARG00000020417 | lysozyme C, tracheal isozyme [Source:NCBI gene;Acc:101102969] | ENSOARG00000020417 | 3 | 150225121 | 150229639 | |
| LYSB | lysozyme C, intestinal isozyme [Source:NCBI gene;Acc:101103222] | ENSOARG00000020429 | 3 | 150266228 | 150270946 | |
| LYZ2 | ENSOARG00000020439 | 3 | 150313875 | 150318870 | ||
| LYZ | lysozyme [Source:NCBI gene;Acc:100049062] | ENSOARG00000020515 | 3 | 150434369 | 150439510 | |
| Cluster3 (2.011) | ENSOARG00000009449 | ENSOARG00000009449 | 10 | 21972591 | 22033480 | |
| PTPN3 | protein tyrosine phosphatase non-receptor type 3 [Source:HGNC Symbol;Acc:HGNC:9655] | ENSOARG00000006968 | 2 | 13790622 | 13898273 | |
| PTPRE | protein tyrosine phosphatase receptor type E [Source:HGNC Symbol;Acc:HGNC:9669] | ENSOARG00000014124 | 22 | 46388779 | 46432003 | |
| PTPN2 | protein tyrosine phosphatase non-receptor type 2 [Source:HGNC Symbol;Acc:HGNC:9650] | ENSOARG00000001930 | 23 | 43434719 | 43479972 | |
| PTPN6 | protein tyrosine phosphatase non-receptor type 6 [Source:HGNC Symbol;Acc:HGNC:9658] | ENSOARG00000005032 | 3 | 207454996 | 207463860 | |
| PPM1B | protein phosphatase, Mg2+/Mn2+ dependent 1B [Source:NCBI gene;Acc:101112467] | ENSOARG00000006389 | 3 | 79989814 | 80014651 | |
| PTPRR | protein tyrosine phosphatase receptor type R [Source:HGNC Symbol;Acc:HGNC:9680] | ENSOARG00000020111 | 3 | 148667786 | 148945673 | |
| PTPRB | protein tyrosine phosphatase receptor type B [Source:HGNC Symbol;Acc:HGNC:9665] | ENSOARG00000020158 | 3 | 148947442 | 149071453 | |
| Cluster4 (2.008) | TMPRSS15 | transmembrane serine protease 15 [Source:HGNC Symbol;Acc:HGNC:9490] | ENSOARG00000015635 | 1 | 136961183 | 137107011 |
| CD163 | CD163 molecule [Source:HGNC Symbol;Acc:HGNC:1631] | ENSOARG00000002862 | 3 | 206525820 | 206562443 | |
| WC1-12 | ENSOARG00000003251 | 3 | 206712983 | 206746704 | ||
| ENSOARG00000003403 | ENSOARG00000003403 | 3 | 206786488 | 206816410 | ||
| ENSOARG00000003988 | ENSOARG00000003988 | 3 | 207068439 | 207075844 | ||
| Cluster5 (1.563) | WDR5B | WD repeat domain 5B [Source:HGNC Symbol;Acc:HGNC:17826] | ENSOARG00000001983 | 1 | 185127233 | 185128225 |
| SEMA5B | semaphorin 5B [Source:HGNC Symbol;Acc:HGNC:10737] | ENSOARG00000020173 | 1 | 185521338 | 185559506 | |
| WDFY2 | WD repeat and FYVE domain containing 2 [Source:HGNC Symbol;Acc:HGNC:20482] | ENSOARG00000008939 | 10 | 21328931 | 21506011 | |
| NBEA | neurobeachin [Source:HGNC Symbol;Acc:HGNC:7648] | ENSOARG00000010627 | 10 | 26007917 | 26592574 | |
| WDR64 | WD repeat domain 64 [Source:HGNC Symbol;Acc:HGNC:26570] | ENSOARG00000007794 | 12 | 33167064 | 33301577 | |
| KCTD3 | ENSOARG00000010181 | 12 | 16935183 | 16992237 | ||
| FBXW2 | F-box and WD repeat domain containing 2 [Source:HGNC Symbol;Acc:HGNC:13608] | ENSOARG00000005690 | 2 | 2693449 | 2722938 | |
| WDR11 | WD repeat domain 11 [Source:HGNC Symbol;Acc:HGNC:13831] | ENSOARG00000004701 | 22 | 39787927 | 39845136 | |
| BUB3 | ENSOARG00000009467 | 22 | 41838447 | 41848859 | ||
| WDR7 | WD repeat domain 7 [Source:HGNCSymbol;Acc:HGNC:13490] | ENSOARG00000005109 | 23 | 56218845 | 56550504 | |
| GNB3 | G protein subunit beta 3 [Source:HGNC Symbol;Acc:HGNC:4400] | ENSOARG00000005728 | 3 | 207555357 | 207560893 | |
| GTF3C4 | general transcription factor IIIC subunit 4 [Source:HGNC Symbol;Acc:HGNC:4667] | ENSOARG00000005793 | 3 | 4112816 | 4127921 | |
| NUP214 | nucleoporin 214 [Source:HGNC Symbol;Acc:HGNC:8064] | ENSOARG00000007056 | 3 | 5440880 | 5534974 | |
| EML4 | EMAP like 4 [Source:HGNC Symbol;Acc:HGNC:1316] | ENSOARG00000007545 | 3 | 81676763 | 81769277 | |
| BIRC6 | baculoviral IAP repeat containing 6 [Source:HGNC Symbol;Acc:HGNC:13516] | ENSOARG00000010515 | 3 | 91252791 | 91461884 | |
| NOL10 | nucleolar protein 10 [Source:HGNC Symbol;Acc:HGNC:25862] | ENSOARG00000015507 | 3 | 19613346 | 19705109 | |
| VPS41 | VPS41 subunit of HOPS complex [Source:HGNC Symbol;Acc:HGNC:12713] | ENSOARG00000017742 | 4 | 82007947 | 82218383 | |
| RACK1 | receptor for activated C kinase 1 [Source:NCBI gene;Acc:100137070] | ENSOARG00000007288 | 5 | 37884620 | 37892060 | |
| HERC1 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 [Source:HGNC Symbol;Acc:HGNC:4867] | ENSOARG00000020775 | 7 | 43237435 | 43432873 | |
| TBC1D31 | TBC1 domain family member 31 [Source:HGNC Symbol;Acc:HGNC:30888] | ENSOARG00000010832 | 9 | 29487020 | 29549424 | |
| Cluster6 (1.419) | ERAP1 | endoplasmic reticulum aminopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:18173] | ENSOARG00000017807 | 5 | 93487749 | 93518183 |
| ERAP2 | endoplasmic reticulum aminopeptidase 2 [Source:HGNC Symbol;Acc:HGNC:29499] | ENSOARG00000017926 | 5 | 93629581 | 93674652 | |
| LNPEP | leucyl and cystinylaminopeptidase [Source:HGNC Symbol;Acc:HGNC:6656] | ENSOARG00000017994 | 5 | 93687872 | 93788740 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez, I.; Fernández, I.; Traoré, A.; Pérez-Pardal, L.; Menéndez-Arias, N.A.; Goyache, F. Ancient Homozygosity Segments in West African Djallonké Sheep Inform on the Genomic Impact of Livestock Adaptation to the Environment. Animals 2020, 10, 1178. https://doi.org/10.3390/ani10071178
Álvarez I, Fernández I, Traoré A, Pérez-Pardal L, Menéndez-Arias NA, Goyache F. Ancient Homozygosity Segments in West African Djallonké Sheep Inform on the Genomic Impact of Livestock Adaptation to the Environment. Animals. 2020; 10(7):1178. https://doi.org/10.3390/ani10071178
Chicago/Turabian StyleÁlvarez, Isabel, Iván Fernández, Amadou Traoré, Lucía Pérez-Pardal, Nuria A. Menéndez-Arias, and Félix Goyache. 2020. "Ancient Homozygosity Segments in West African Djallonké Sheep Inform on the Genomic Impact of Livestock Adaptation to the Environment" Animals 10, no. 7: 1178. https://doi.org/10.3390/ani10071178
APA StyleÁlvarez, I., Fernández, I., Traoré, A., Pérez-Pardal, L., Menéndez-Arias, N. A., & Goyache, F. (2020). Ancient Homozygosity Segments in West African Djallonké Sheep Inform on the Genomic Impact of Livestock Adaptation to the Environment. Animals, 10(7), 1178. https://doi.org/10.3390/ani10071178







