Validation of Real-time PCR Reference Genes of Muscle Metabolism in Harvested Spiny-Cheek Crayfish (Faxonius limosus) Exposed to Seasonal Variation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Total RNA Isolation and cDNA Synthesis
2.3. Identification and Characterization of Candidate and Target Genes
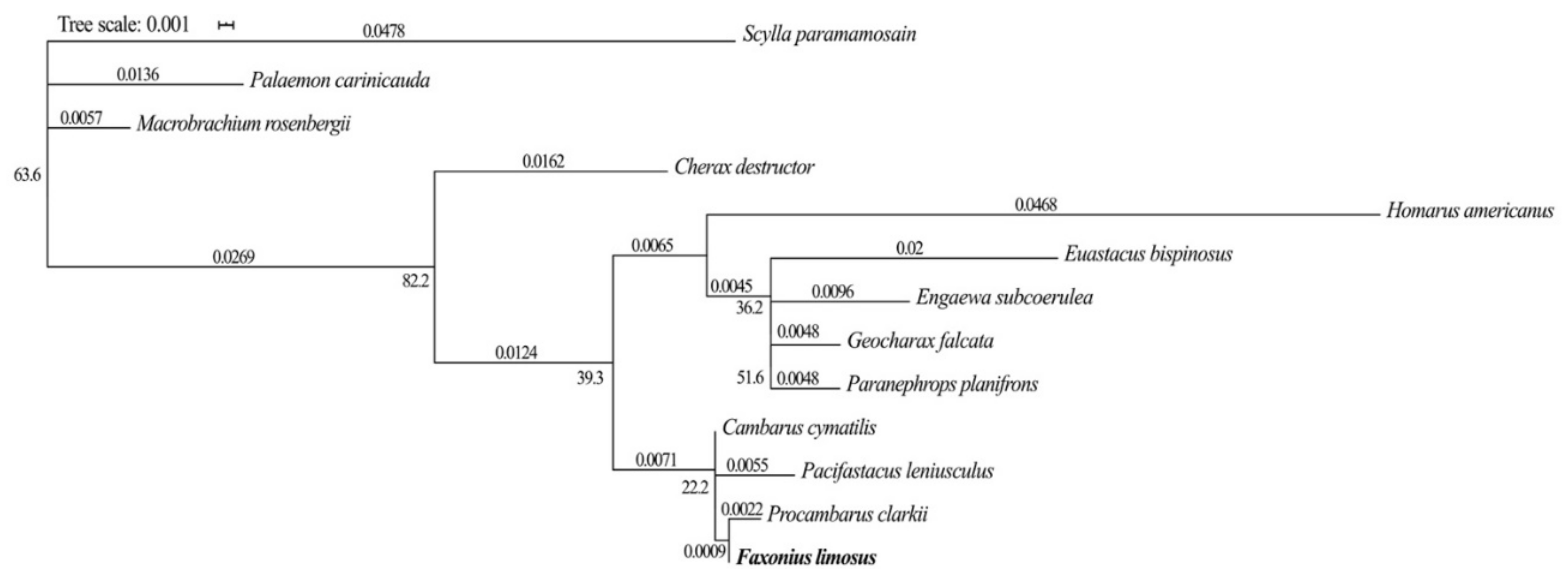
2.4. Phylogenetic Analysis of the Gapdh Gene
2.5. Quantitative Real-Time PCR
2.6. Algorithms for the Selection of Reference Genes
2.7. Gene Expression Profiling
3. Results and Discussion
3.1. Sequences and Phylogenetic Analysis of the Candidate and Target Genes
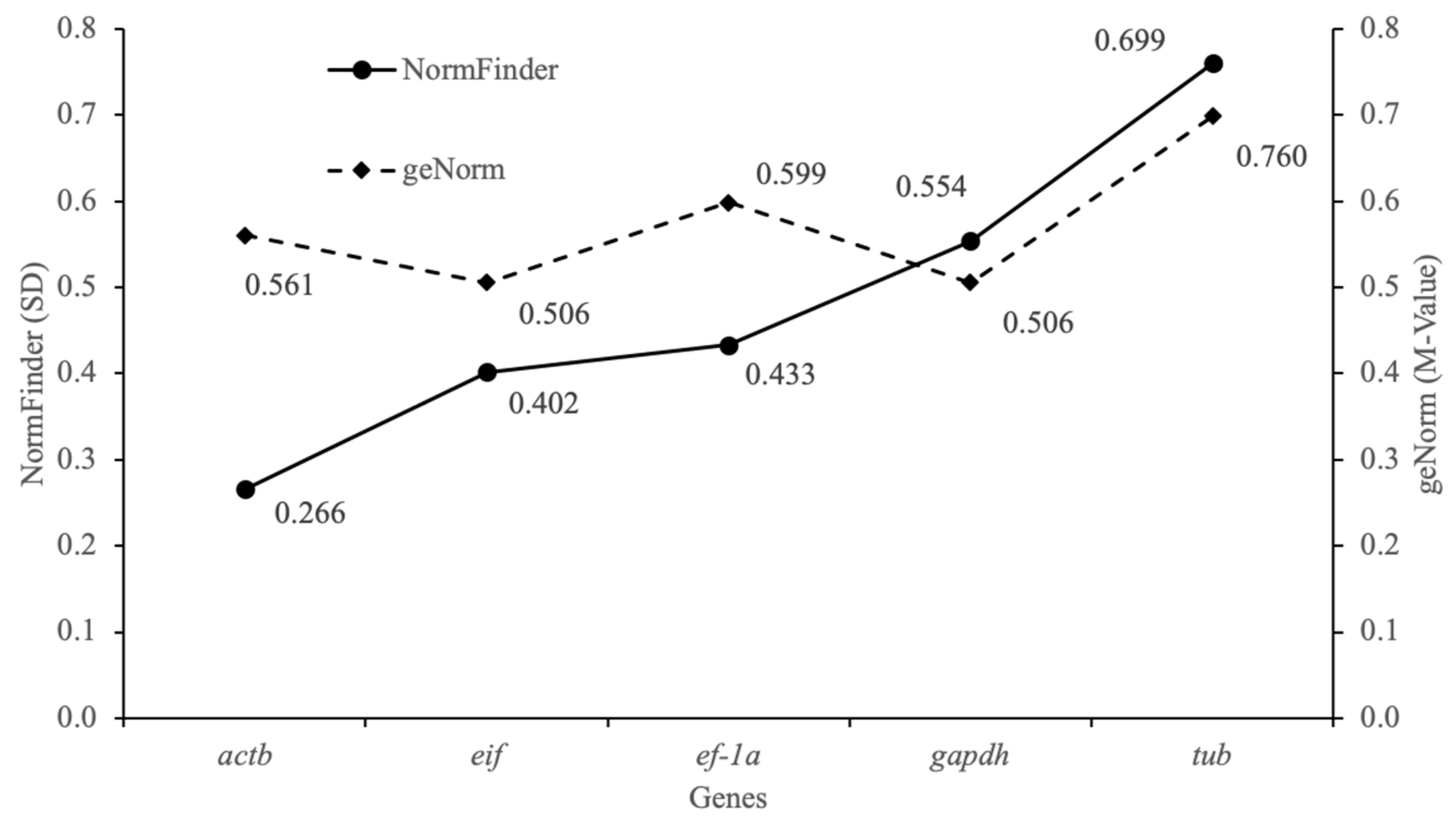
3.2. Stability and Ranking of Reference Genes in F. limosus
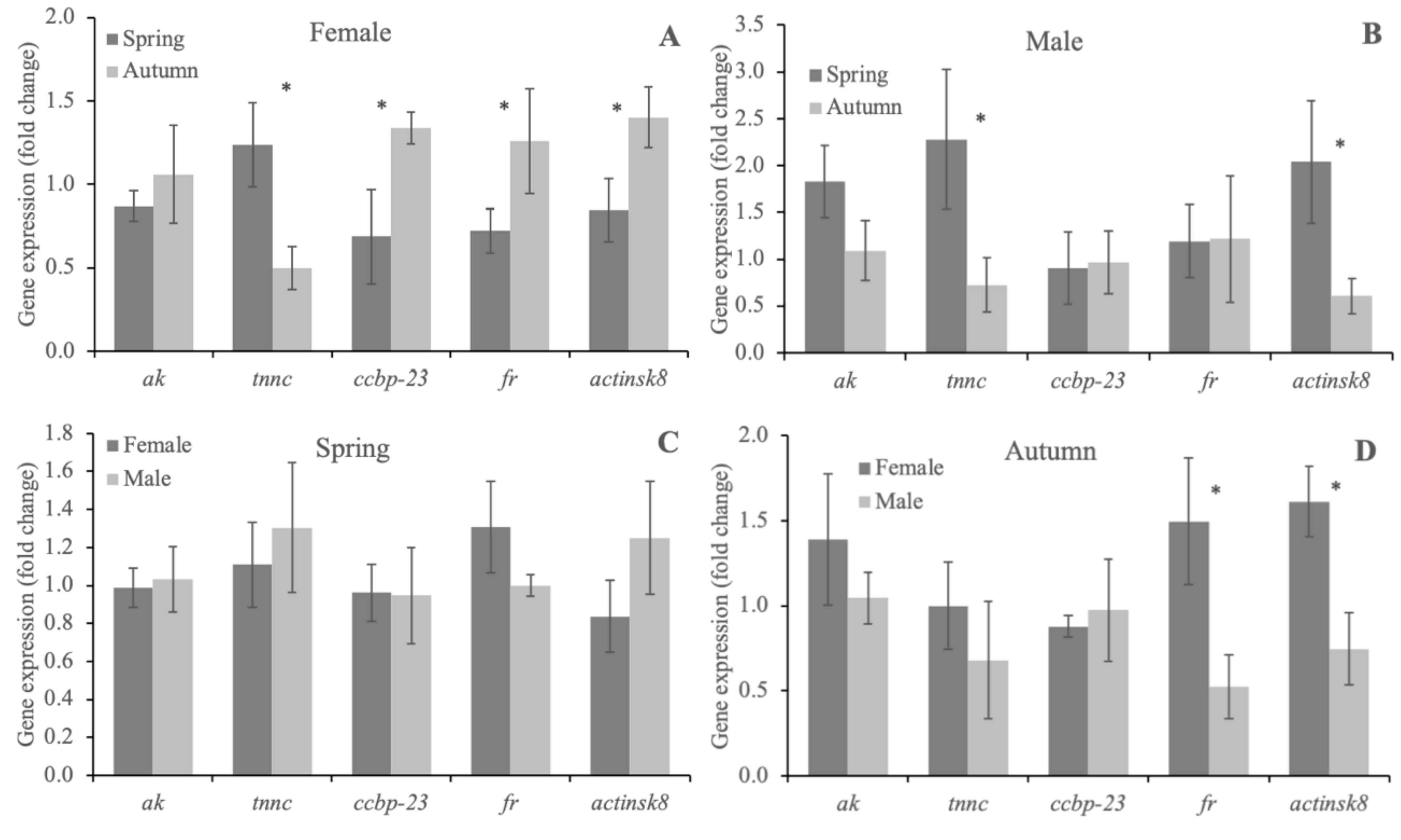
3.3. Quantitative Analysis of ak, tnnc, ccbp-23, fr and actinsk8 in the Abdomen Muscles of Females and Males of F. limosus from Two Seasons
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stanek, M.; Borejszo, Z.; Dąbrowski, J.; Janicki, B. Fat and cholesterol content and fatty acid profiles in edible tissues of spiny-cheek crayfish (Orconectes limosus Raf.) from Lake Gopło (Poland). Arch. Pol. Fish. 2011, 19, 241–278. [Google Scholar] [CrossRef]
- Fonseca, L.F.S.; Gimenez, D.F.J.; Dos Santos Silva, D.B.; Barthelson, R.; Baldi, F.; Ferro, J.A.; Albuquerque, L.G. Differences in global gene expression in muscle tissue of Nellore cattle with divergent meat tenderness. BMC Genom. 2017, 18, 945. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, S.; Panguluri, S.K.; Kumar, A. Gene profiling studies in skeletal muscle by quantitative real-time polymerase chain reaction assay. In Myogenesis; Humana Press: Totowa, NJ, USA, 2012; pp. 311–324. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Qian, Z.; Lu, W.; Ding, H.; Yu, H.; Wang, H.; Li, J. Identification and Characterization of Reference Genes for Normalizing Expression Data from Red Swamp Crawfish Procambarus clarkii. Int. J. Mol. Sci. 2015, 16, 21591–21605. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Han, S.; Fei, J.; Zhang, L.; Ray, J.W.; Wang, W.; Li, Y. Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii. Genes 2019, 10, 645. [Google Scholar] [CrossRef] [Green Version]
- Decker, E.A.; Welch, B. Role of ferritin as a lipid oxidation catalyst in muscle food. J. Agric. Food Chem. 1990, 38, 674–677. [Google Scholar] [CrossRef]
- Dawson, N.J.; Storey, K.B. Regulation of tail muscle arginine kinase by reversible phosphorylation in an anoxia-tolerant crayfish. J. Comp. Physiol. 2011, 181, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, N.M.; Taylor, E.W.; El Haj, A.J. Seasonal and latitudinal adaptation to temperature in crustaceans. J. Therm. Biol. 1997, 22, 419–427. [Google Scholar] [CrossRef]
- Gills, T.; Marshall, R.; Tibbits, G. Functional and evolutionary relationships of troponin C. Physiol. Genom. 2007, 32, 16–27. [Google Scholar] [CrossRef]
- Macabelli, C.H.; Ferreira, R.M.; Gimenes, L.U.; de Carvalho, N.A.T.; Soares, J.G.; Ayres, H.; Ferraz, M.L.; Watanabe, Y.F.; Watanabe, O.Y.; Sangalli, J.R.; et al. Reference gene selection for gene expression analysis of oocytes collected from dairy cattle and buffaloes during winter and summer. PLoS ONE 2014, 9, e93287. [Google Scholar] [CrossRef] [Green Version]
- Panicz, R. Validation of reference genes for RT-qPCR analysis of growth hormone receptor and growth hormone expression in the tench (Tinca tinca) fed substituting poultry meal for fish meal. Aquaculture 2016, 465, 179–188. [Google Scholar] [CrossRef]
- Strużyński, W. Raki w wodach Polski–ich dramaty i kariery. Salamandra Mag. Przyr. 2001, 14, 1. (in Polish). [Google Scholar]
- Śmietana, P. Uwarunkowania Rozmieszczenia i Mechanizmy Konkurencji Międzygatunkowej Raka Szlachetnego (Astacus astacus L.) i Raka Pręgowatego (Orconectes limosus Raf.); Wydawnictwo Naukowe Uniwersytetu Szczecińskiego: Polska, Poland, 2013; pp. 5–266. (in Polish) [Google Scholar]
- Dąbrowski, T.; Kołakowski, E.; Wawreszuk, H.; Choroszucha, C. Studies on chemical composition of American crayfish (Orconectes limosus) meat as related to its nutritive value. J. Fish. Board Can. 1966, 23, 1653–1662. [Google Scholar] [CrossRef]
- Stanek, M.; Kupcewicz, B.; Dąbrowski, J.; Janicki, B. Ocena zawartości tłuszczu i profilu kwasów tłuszczowych w mięsie raka pręgowatego (Orconestes limosus Raf.) z rzeki Brdy i jeziora Gopło. J. Cent. Eur. Agric. 2010, 11, 297–304. [Google Scholar]
- Choubert, G.; Mendes-Pinto, M.M.; Morais, R. Pigmenting efficacy of astaxanthin fed to rainbow trout Oncorhynchus mykiss: Effect of dietary astaxanthin and lipid sources. Aquaculture 2006, 257, 429–436. [Google Scholar] [CrossRef]
- Struszczyk, M.H. Chitin and Chitosan. Part II Applications of Chitosan. Polimery 2002, 47, 396–403. [Google Scholar] [CrossRef]
- Shen, H.; Hu, Y.; Ma, Y.; Zhou, X.; Shui, Y.; Li, C.; Xu, P.; Sun, X. In-Depth Transcriptome Analysis of the Red Swamp Crayfish Procambarus clarkii. PLoS ONE 2014, 9, e110548. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformm. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR; Meuer, S., Wittwer, C., Nakagawara, K., Eds.; Methods and Applications, Springer Press: Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Vandesompele, J.; De Preter, F.; Pattyn, K.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.T.; Elder Jr, J.F.; Thompson, S.M.; Hightower, P.; Bechler, D. Phylogeny of the freshwater crayfish subfamily Cambarinae based on 16S rDNA gene analysis. Res. Trends 2011, 2, 97–113. [Google Scholar]
- Crandall, K.A.; De Grave, S. An updated classification of the freshwater crayfishes (Decapoda: Astacidea) of the world, with a complete species list. J. Crustacean Biol. 2017, 37, 615–653. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Sun, M.; Liu, X. Phase-specific expression of an insulin-like androgenic gland factor in a marine shrimp Lysmata wurdemanni: Implication for maintaining protandric simultaneous hermaphroditism. PLoS ONE 2017, 12, e0172782. [Google Scholar] [CrossRef]
- Reyes, A.; Salazar, M.; Granja, C. Temperature modifies gene expression in subcuticular epithelial cells of white spot syndrome virus-infected Litopenaeus vannamei. Dev. Comp. Immunol. 2007, 31, 23–29. [Google Scholar] [CrossRef]
- Liu, S.; Qi, C.; Jia, Y.; Gu, Z.; Li, E. Growth and intestinal health of the red claw crayfish, Cherax quadricarinatus, reared under different salinities. Aquaculture 2020, 524, 735256. [Google Scholar] [CrossRef]
- Jiang, H.; Xing, Z.; Lu, W.; Qian, Z.; Yu, H.; Li, J. Transcriptome Analysis of Red Swamp Crawfish Procambarus clarkii Reveals Genes Involved in Gonadal Development. PLoS ONE 2014, 9, e105122. [Google Scholar] [CrossRef] [Green Version]
- Ergin, B.; Purali, N. Comparison of the efficiency and relevancy of reference genes in crayfish tissue samples/Kerevit doku örneklerinde referans genlerin geçerlilik ve etkinliğinin karşılaştırılması. Turk. J. Biochem. 2016, 41, 144–149. [Google Scholar] [CrossRef]
- Nicosia, A.; Salvatore, C.; Tagliavia, M.; Maggio, T.; Salamone, M.; Adamo, G.; Ragusa, M.A.; Bennici, C.; Masullo, T.; Mazzola, S.; et al. The nucleic acid-binding protein PcCNBP is transcriptionally regulated during the immune response in red swamp crayfish Procambarus clarkii. Cell Stress Chaperones 2016, 21, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Leelatanawit, R.; Klanchui, A.; Uawisetwathana, U.; Karoonuthaisiri, N. Validation of reference genes for real-time PCR of reproductive system in the black Tiger shrimp. PLoS ONE 2012, 7, e52677. [Google Scholar] [CrossRef] [Green Version]
- Gatzmann, F.; Falckenhayn, C.; Gutekunst, J.; Hanna, K.; Raddatz, G.; Carneiro, V.C.; Lyko, F. The methylome of the marbled crayfish links gene body methylation to stable expression of poorly accessible genes. Epigenetics Chromatin 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhar, A.K.; Bowers, R.M.; Licon, K.S.; Veazey, G.; Read, B. Validation of reference genes for quantitative measurement of immune gene expression in shrimp. Mol. Immunol. 2009, 46, 1688–1695. [Google Scholar] [CrossRef] [PubMed]
- Haiech, J.; Moreau, M.; Leclerc, C.; Kilhoffer, M.C. Facts and conjectures on calmodulin and its cousin proteins, parvalbumin and troponin C. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol. Rev. 1996, 76, 371–423. [Google Scholar] [CrossRef]
- Chao, E.; Kim, H.W.; Mykles, D.L. Cloning and tissue expression of eleven troponin-C isoforms in the American lobster, Homarus americanus. Comp. Biochem. Physiol. B 2010, 157, 88–101. [Google Scholar] [CrossRef]
- Suzuki, Y. Abdominal abductor muscle in crayfish: Physiological properties and neural control. I-twitch system. Comp. Biochem. Physiol. A Physiol. 1998, 59, 249–258. [Google Scholar] [CrossRef]
- MacLea, K.S.; Covi, J.A.; Kim, H.W.; Chao, E.; Medler, S.; Chang, E.S.; Mykles, D.L. Myostatin from the American lobster, Homarus americanus: Cloning and effects of molting on expression in skeletal muscles. Comp. Biochem. Phys. A 2010, 157, 328–337. [Google Scholar] [CrossRef]
- Carter, T.J.; Fraser, P.J. Effect of temperature on tilt evoked swimming in the crabs Carcinus and Macropipus. J. Therm. Biol. 1991, 16, 367–375. [Google Scholar] [CrossRef]
- Hartnoll, R.G. Growth. In The Biology of Crustacea, 2nd ed.; Abele, L.G., Ed.; Academic Press: New York, NY, USA, 1982; pp. 111–196. [Google Scholar]
- Medler, S.; Brown, K.J.; Chang, E.S.; Mykles, D.L. Eyestalk ablation has little effect on actin and myosin heavy chain gene expression in adult lobster skeletal muscles. Biol. Bull. 2005, 208, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Sauter, A.; Staudenmann, W.; Hughes, G.J.; Heizmann, C.W. A novel EF-hand Ca2+-binding protein from abdominal muscle of crustaceans with similarity to calcyphosine from dog thyroidea. Eur. J. Biochem. 1995, 227, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, P.D.; Bentley, M.G.; Watson, G.J. Mating behaviour and evidence for a female released courtship pheromone in the signal crayfish Pacifastacus leniusculus. J. Chem. Ecol. 2003, 29, 465–475. [Google Scholar] [CrossRef]
- Durand, J.P.; Goudard, F.; Pieri, J.; Escoubas, J.M.; Schreiber, N.; Cadoret, J.P. Crassostrea gigas ferritin: cDNA sequence analysis for two heavy chain type subunits and protein purification. Gene 2004, 338, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Le, N.T.V.; Richarson, D.E. The role of iron in cell cycle progression and the proliferation of neoplastic cells. Biophys. Biochim. Acta 2003, 1603, 31–46. [Google Scholar] [CrossRef]
- Govind, C.K.; She, J.; Lang, F. Lengthening of lobster muscle fibers by two age-dependent mechanisms. Experientia 1977, 33, 35–36. [Google Scholar] [CrossRef]
- Mittenthal, J.E.; Olson, M.C.; Cummings, G.D. Morphology of the closer muscles in normal and homoeotic legs of crayfish. Biol. Bull. 1980, 159, 714–727. [Google Scholar] [CrossRef]
- Houlihan, D.F.; El Haj, A.J. An analysis of muscle growth. In Crustacean Issues 3: Factors in Adult Growth; Wenner, A., Ed.; A.A. Balkema Press: Rotterdam, the Netherlands, 2003; pp. 15–29. [Google Scholar]
- Stephens, P.J.; Mellon, D. Modification of structure and physiology in transformed shrimp muscle. J. Comp. Physiol. 1979, 132, 97–108. [Google Scholar] [CrossRef]
- Shofer, S.L.; Willis, J.A.; Tjeerdema, R.S. Effects of hypoxia and toxicant exposure on arginine kinase function as measures by 31P-NMR magnetization transfer in living abalone. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1997, 117, 283–289. [Google Scholar] [CrossRef]
- Yin, S.J.; Zhang, L.; Zhang, L.; Wan, J.; Song, W.; Jiang, X.; Yong-Doo, P.; Si, Y.X. Metabolic responses and arginine kinase expression of juvenile cuttlefish (Sepia pharaonis) under salinity stress. Int. J. Biol. Macromol. 2018, 113, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Di, G.; Kong, X.; Zhu, G.; Liu, S.; Zhang, C.; Ke, C. Pathology and physiology of Haliotis diversicolor with withering syndrome. Aquaculture 2016, 453, 1–9. [Google Scholar] [CrossRef]
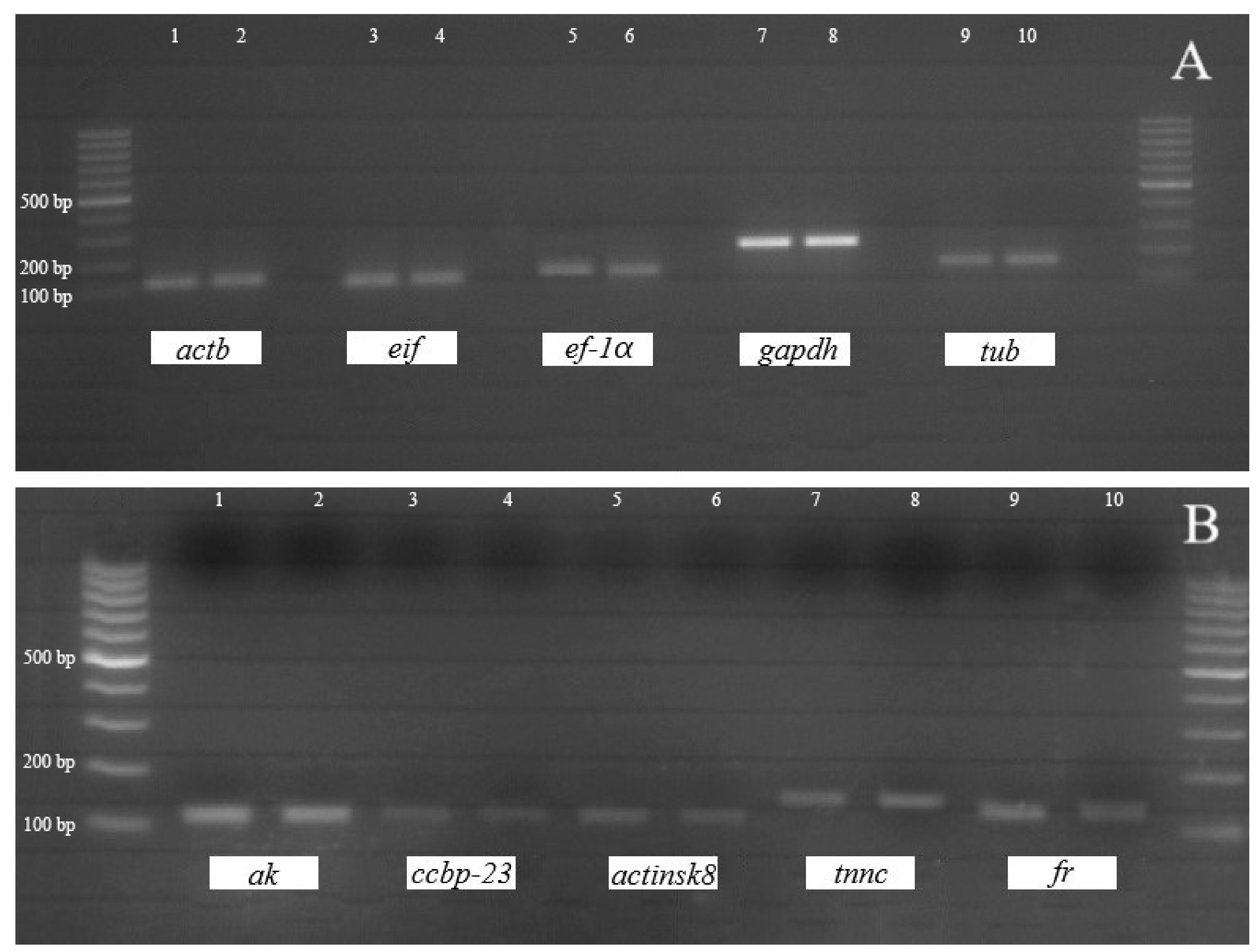
| Gene Name (Symbol) | Cellular Function | Primer Sequence (5′ → 3′) | Product Size(bp) | Reference |
|---|---|---|---|---|
| α-tubulin (α-tub) | Cytoskeletal structural protein | F: CCACTTCCCTCTCGTCACC R: AACAGCACGCCATGTACTTT | 155 | [4] |
| β-actin (actb) | Cytoskeletal structural protein | F: ATTGCAGACAGGATGCAGAA R: GAAAGGGAAGCCAAGATGG | 125 | |
| Elongation factor 1-α (eif-1a) | Protein biosynthesis | F: CCACAAAGGCAGGTGAAAAGG R: ATTGGGTGAACCAAGCAGGG | 110 | |
| Eukaryotic translation initiation factor 5a (eif) | Protein synthesis | F: GGAATAAGGGGACGAAGACC R: GCAAACACACGCTGGGAT | 126 | |
| Glyceraldehyde-3-phosphate dehydrogenase (gapdh) | Oxidoreductase in glycolysis and gluconeogenesis | F: GCCCAGAACATCATCCCATCT R: CGTCATCCTCAGTGTAACCCAAG | 235 | |
| Arginine kinase (ak) | High-energy phosphate transfer | F: GGAGTGAGTCAGTGTGAGGC R: GGAGTAGCGTTACCCACGAG | 114 | [18] |
| Crustacean calcium-binding protein 23 (ccbp-23) | Regulatory protein | F: GGCCATCGATGAAGACGAGT R: TCCGGATCAGCAGTAGTGGA | 117 | |
| Ferritin (fr) | Iron binding | F: GCTCTTCCTGGGGCATCAAA R: CAGTGTTGGTGCTGCAATGG | 132 | |
| Skeletal muscle actin 8 (actinsk8) | Myofibrillar contraction protein | F: CCCGCCACTTACGTTACCAT R: ACCTTCATAGACGGGGACCA | 117 | |
| Troponin C (tnnc) | Conformational changes on the thin filament | F: TCAGGAGGTCATCGCTGAGA R: GCCCTTGTCATAGATGCGGA | 151 |
| FAS 1 | MAS 2 | FAA 3 | MAA 4 | |||||
|---|---|---|---|---|---|---|---|---|
| FAS 1 | 0.301 | 0.432 | 0.150 | 0.223 | 0.211 | 0.284 | ||
| MAS 2 | eif | actb | 0.341 | 0.238 | 0.416 | 0.404 | ||
| FAA 3 | eif | actb | actb | actb | 0.132 | 0.168 | ||
| MAA 4 | eif | actb | actb | actb | eif | actb | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Śmietana, N.; Panicz, R.; Sobczak, M.; Eljasik, P.; Śmietana, P. Validation of Real-time PCR Reference Genes of Muscle Metabolism in Harvested Spiny-Cheek Crayfish (Faxonius limosus) Exposed to Seasonal Variation. Animals 2020, 10, 1140. https://doi.org/10.3390/ani10071140
Śmietana N, Panicz R, Sobczak M, Eljasik P, Śmietana P. Validation of Real-time PCR Reference Genes of Muscle Metabolism in Harvested Spiny-Cheek Crayfish (Faxonius limosus) Exposed to Seasonal Variation. Animals. 2020; 10(7):1140. https://doi.org/10.3390/ani10071140
Chicago/Turabian StyleŚmietana, Natalia, Remigiusz Panicz, Małgorzata Sobczak, Piotr Eljasik, and Przemysław Śmietana. 2020. "Validation of Real-time PCR Reference Genes of Muscle Metabolism in Harvested Spiny-Cheek Crayfish (Faxonius limosus) Exposed to Seasonal Variation" Animals 10, no. 7: 1140. https://doi.org/10.3390/ani10071140
APA StyleŚmietana, N., Panicz, R., Sobczak, M., Eljasik, P., & Śmietana, P. (2020). Validation of Real-time PCR Reference Genes of Muscle Metabolism in Harvested Spiny-Cheek Crayfish (Faxonius limosus) Exposed to Seasonal Variation. Animals, 10(7), 1140. https://doi.org/10.3390/ani10071140






