Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Isolation
2.2. Microsatellite Genotyping
2.3. Statistical Analysis
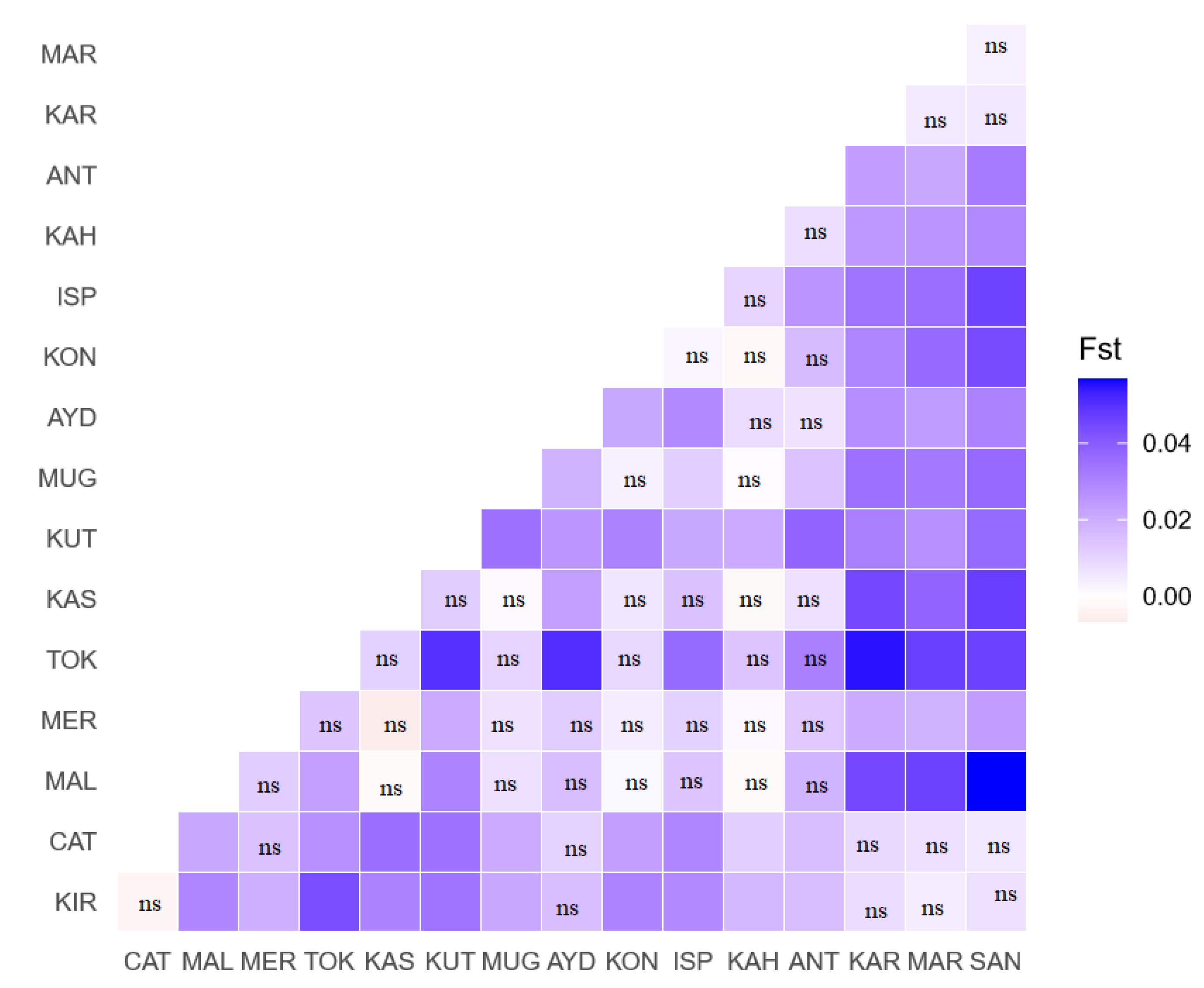
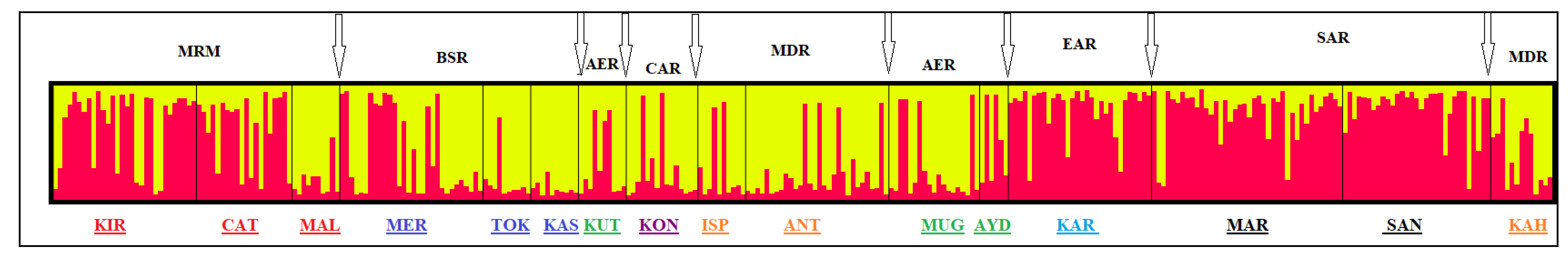
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mittermeier, R.A.; Gil, P.R.; Hoffman, M.; Pilgrim, J.; Brooks, T.; Mittermeier, C.G. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. Conserv. Int. 2005, 392. [Google Scholar]
- Şekercioğlu, C.H.; Anderson, S.; Akcay, E.; Bilgin, R.; Can, O.E.; Semiz, G.; Tavşanoğlu, C.; Yokeş, M.B.; Soyumert, A.; Ipekdal, K.; et al. Turkey’s globally important biodiversity in crisis. Boil. Conserv. 2011, 144, 2752–2769. [Google Scholar] [CrossRef]
- Kugler, W.; Grünenfelder, H.-P.; Broxham, E. Donkey Breeds in Europe. Inventory, Description, Need for Action: Report. Nat. Sustain. 2018, 679–685. [Google Scholar]
- Yilmaz, O.; Ertuğrul, M. Domestication of Donkey (Equus Asinus). Iğdır Üniversitesi Fen Bilim. Enstitüsü Derg. 2011, 1, 111–115. [Google Scholar]
- Yılmaz, O.; Wilson, T.R. The Domestic Livestock Resources of Turkey: Notes on Donkeys. J. Anim. Plant. Sci. 2013, 23, 651–656. [Google Scholar]
- Güleç, E. Eşek Yetiştiriciliği ve Önemi; Bilgi Müşavirlik ve Mühendislik: Ankara, Turkey, 2010; p. 317. [Google Scholar]
- Molecular Genetic Characterization of Animal Genetic Resources (FAO). In FAO Animal Production and Health Guidlines. No. 9; FAO: Rome, Italy, 2011.
- Guérin, G.; Nocart, M.; Kemp, S.J. Fifteen new synteny assignments of microsatellites to the bovine genome. Anim. Genet. 2009, 25, 179–181. [Google Scholar] [CrossRef]
- Irvin, Z.; Giffard, J.; Brandon, R.; Breen, M.; Bell, K. Equine dinucleotide repeat polymorphisms at loci ASB 21, 23, 25 and 37-43. Anim. Genet. 1998, 29, 67. [Google Scholar]
- Marklund, S.; Ellegren, H.; Eriksson, S.; Sandberg, K.; Andersson, L. Parentage testing and linkage analysis in the horse using a set of highly polymorphic microsatellites. Anim. Genet. 2009, 25, 19–23. [Google Scholar] [CrossRef]
- Binns, M.; Holmes, N.G.; Rolliman, A.; Scott, A. The identification of polymorphicmicrosatellite loci in the horse and their use in thoroughbred parentage testing. Br. Veter J. 1995, 151, 9–15. [Google Scholar] [CrossRef]
- Ruth, L.S.; Hopman, T.J.; Schug, M.D.; Aquadro, C.F.; Bowling, A.T.; Murray, J.D.; Caetano, A.R.; Antczak, D.F. Equine dinucleotide repeat loci COR041-COR060. Anim. Genet. 1999, 30, 320–321. [Google Scholar] [CrossRef]
- van Haeringen, W.A.; van de Goor, L.H.; van der Hout, N.; Lenstra, J.A. Characterization of 24 equine microsatellite loci. Anim. Genet. 1998, 29, 153–156. [Google Scholar] [PubMed]
- Breen, M.; Lindgren, G.; Binns, M.M.; Norman, J.; Irvin, Z.; Bell, K.; Sandberg, K.; Ellegren, H. Genetical and physical assignments of equine microsatellites-first integration of anchored markers in horse genome mapping. Mamm. Genome 1997, 8, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Guérin, G.; Bertaud, M. Characterization of two polymorphic horse microsatellites: HMS15 and HMS20. Anim. Genet. 1996, 27, 123. [Google Scholar] [PubMed]
- Hopman, T.J.; Han, E.B.; Story, M.R.; Bowling, A.T.; Caetano, A.R.; Antczak, D.F.; Schug, M.D.; Aquadro, C.F.; Murray, J.D. Equine dinucleotide repeat loci COR001-COR020. Anim. Genet. 1999, 30, 225–226. [Google Scholar] [CrossRef] [PubMed]
- Coogle, L.; Bailey, E. Equine dinucleotide repeat loci LEX049-LEX063. Anim. Genet. 1997, 28, 378. [Google Scholar] [CrossRef]
- Ellegren, H.; Johansson, M.; Sandberg, K.; Andersson, L. Cloning of highly polymorphic microsatellites in the horse. Anim. Genet. 2009, 23, 133–142. [Google Scholar] [CrossRef]
- Tallmadge, R.L.; Hopman, T.J.; Schug, M.D.; Aquadro, C.F.; Bowling, A.T.; Murray, J.D.; Caetano, A.R.; Antczak, D.F. Equine dinucleotide repeat loci cor061-cor080. Anim. Genet. 1999, 30, 462–466. [Google Scholar] [CrossRef]
- Tallmadge, R.L.; Evans, K.G.; Hopman, T.J.; Schug, M.D.; Aquadro, C.F.; Bowling, A.T.; Murray, J.D.; Caetano, A.R.; Antczak, D.F. Equine dinucleotide repeat loci COR081-COR100. Anim. Genet. 1999, 30, 470. [Google Scholar] [CrossRef]
- Belkhir, K.B.P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel Sous Windows TM Pour La Genetique Des. Populations; CNRS UMR 5000, Université de Montpellier II: Montpellier, France, 1996. [Google Scholar]
- Goudet, J. FSTAT (Version 2.9.4). In Department of Ecology & Evolution, Biology Building; UNIL: Lausanne, Switzerland; Available online: https://www2.unil.ch/popgen/softwares/fstat.htm10.1017/CBO9781107415324.004 (accessed on 21 May 2020).
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE Version 1.31: Microsoft Window-Based Freeware for Population Genetics Analysis: Quick User Guide. University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2007, 1, 47–50. [Google Scholar] [CrossRef]
- Nei, M. Estimation of Average Heterozygosity and Genetic Distance from a Small Number of Individuals. Genetics 1978, 89, 583–590. [Google Scholar] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Boil. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.; Vonholdt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2003, 4, 137–138. [Google Scholar] [CrossRef]
- Stanisic, L.; Dimitrijevic, V.; Simeunovic, P.; Glavinic, U.; Jovanovic, B.; Stevanovic, J.; Stanimirović, Z. Assessment of 17 microsatellite loci for their use in parentage verification and individual identification in the Balkan donkey breed. Genetics 2017, 49, 21–30. [Google Scholar] [CrossRef]
- Jordana, J.; Gómez, M.; Aranguren-Méndez, J. Genetic diversity in Spanish donkey breeds using microsatellite DNA markers. Genet. Sel. Evol. 2001, 33, 433–442. [Google Scholar] [CrossRef]
- Jordana, J.; Folch, P.; Aranguren, J.A. Microsatellite analysis of genetic diversity in the Catalonian donkey breed. J. Anim. Breed. Genet. 2001, 118, 57–63. [Google Scholar] [CrossRef]
- Ivankovic, A.; Kavar, T.; Caput, P.; Mioc, B.; Pavic, V.; Dovc, P. Genetic diversity of three donkey populations in the Croatian coastal region. Anim. Genet. 2002, 33, 169–177. [Google Scholar] [CrossRef]
- Zhu, W.; Su, Y.; Liu, Y.; Ni, J.; Wu, J. Microsatellite Polymorphism Analysis of Yang Yuan Donkey in China. J. Anim. Vet. Adv. 2013, 12, 795–797. [Google Scholar]
- Zhang, R.; Xie, W.; Zhang, T.; Lei, C. High polymorphism at microsatellite loci in the Chinese donkey. Genet. Mol. Res. 2016, 15, 15028291. [Google Scholar] [CrossRef] [PubMed]
- Di, R.; Liu, Q.; Xie, F.; Hu, W.; Wang, X.-Y.; Cao, X.-H.; Pan, Z.-Y.; Chen, G.; Chu, M. Evaluation of genetic diversity and population structure of five Chinese indigenous donkey breeds using microsatellite markers. Czech J. Anim. Sci. 2017, 62, 219–225. [Google Scholar] [CrossRef]
- Zeng, L.; Dang, R.; Dong, H.; Li, F.; Chen, H.; Lei, C. Genetic diversity and relationships of Chinese donkeys using microsatellite markers. Arch. Anim. Breed. 2019, 62, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Aroua, M.; Jemmali, B.; Ouled Ahmed, H.; Khaldi, S.; Ben Said, S.; Baccouche, R.; Mahouachi, M. Genetic Characterization of Tunisian Donkey with DNA Microsatellites. Ind. J. Anim. Sci. 2019, 89, 966–969. [Google Scholar]
- Ciampolini, R.; Cecchi, F.; Mazzanti, E.; Ciani, E.; Tancredi, M.; de Sanctis, B. The genetic variability analysis of the Amiata donkey breed by molecular data. Ital. J. Anim. Sci. 2007, 6, 78–80. [Google Scholar] [CrossRef]
- Colli, L.; Perrotta, G.; Negrini, R.; Bomba, L.; Bigi, D.; Zambonelli, P.; Supplizi, A.V.; Liotta, L.; Marsan, P.A. Detecting population structure and recent demographic history in endangered livestock breeds: The case of the Italian autochthonous donkeys. Anim. Genet. 2012, 44, 69–78. [Google Scholar] [CrossRef]
- Bordonaro, S.; Guastella, A.M.; Criscione, A.; Zuccaro, A.; Marletta, D. Genetic Diversity and Variability in Endangered Pantesco and Two Other Sicilian Donkey Breeds Assessed by Microsatellite Markers. Sci. World J. 2012, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Matassino, D.; Cecchi, F.; Ciani, F.; Incoronato, C.; Occidente, M.; Santoro, L.; Ciampolini, R. Genetic Diversity and Variability in two Italian Autochthonous Donkey Genetic Types Assessed by Microsatellite Markers. Ital. J. Anim. Sci. 2014, 13, 3028. [Google Scholar] [CrossRef]
- Drummond, A.; Alves, E.D. Perfil socioeconômico e demográfico e a capacidade funcional de idosos atendidos pela Estratégia Saúde da Família de Paranoá, Distrito Federal. Rev. Bras. Geriatr. Gerontol. 2013, 16, 727–738. [Google Scholar] [CrossRef]
- Jordana, J.; Ferrando, A.; Miró, J.; Goyache, F.; Loarca, A.; López, O.M.; Canelon, J.; Stemmer, A.; Aguirre, L.; Lara, M.; et al. Genetic relationships among American donkey populations: Insights into the process of colonization. J. Anim. Breed. Genet. 2015, 133, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Dakin, E.E.; Avise, J.C. Microsatellite null alleles in parentage analysis. Heredity 2004, 93, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B.D.; Hamattal, M.A.; Touré, F.A.; Nantoumé, A.D. Watermelons in the Sand of Sahara: Cultivation and use of indigenous landraces in the Tombouctou Region of Mali. Ethnobot. Res. Appl. 2011, 9, 151–162. [Google Scholar] [CrossRef]
- Seyedabadi, H.; Amirinia, C.; Banabazi, M.H.; Emrani, H. Parentage Verification of Iranian Caspian Horse Using Microsatellites Markers. Iran. J. Biotechnol. 2006, 4, 260–264. [Google Scholar]
- Rosenbom, S.; Costa, V.; Al-Araimi, N.A.; Kefena, E.; Abdel-Moneim, A.S.; Abdalla, M.A.; Bakhiet, A.; Beja-Pereira, A. Genetic diversity of donkey populations from the putative centers of domestication. Anim. Genet. 2014, 46, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, M.C.; Valiati, P.; Cherchi, R.; Gorla, E.; Prinsen, R.T.M.M.; Longeri, M.; Bagnato, A.; Strillacci, M.G. Mitochondrial DNA genetic diversity in six Italian donkey breeds (Equus asinus). Mitochondrial DNA Part A 2017, 29, 409–418. [Google Scholar] [CrossRef]
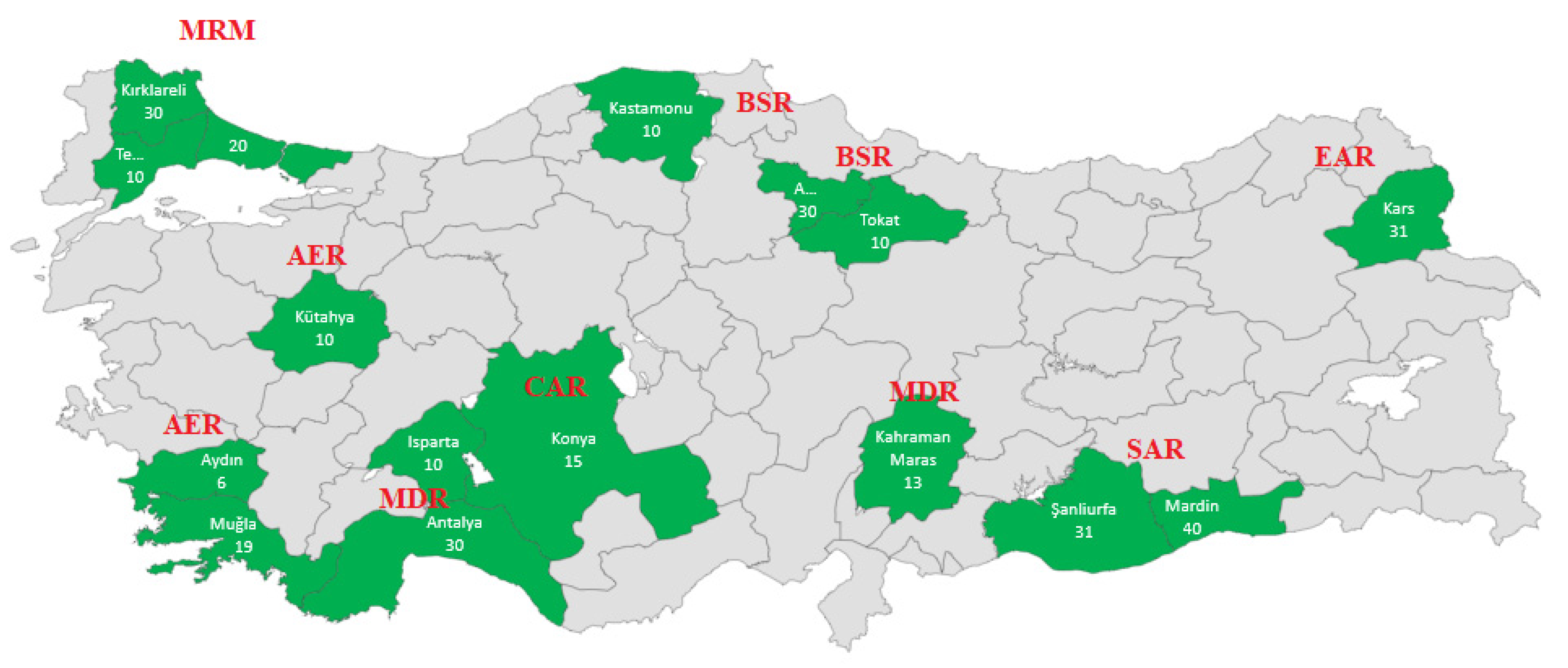

| Region | Locations | Geographical Location | Number of Samples | |
|---|---|---|---|---|
| Marmara (MRM) | Kırklareli | 41°51′N | 27°19′E | 30 |
| İstanbul-Çatalca | 41°06′N | 28°30′E | 20 | |
| Tekirdağ-Malkara | 40°52′N | 26°57′E | 10 | |
| Black Sea (BSR) | Amasya-Merzifon | 40°53′N | 35°32′E | 30 |
| Tokat | 40°12′N | 36°27′E | 10 | |
| Kastamonu-Cide | 41°50′N | 32°54′E | 10 | |
| Aegean (AER) | Kütahya | 39°21′N | 30°01′E | 10 |
| Muğla | 36°37′N | 29°26′E | 19 | |
| Aydın | 37°44′N | 28°01′E | 6 | |
| Central Anatolia (CAR) | Konya | 37°38′N | 32°26′E | 15 |
| Mediterranean (MDR) | Isparta | 37°49′N | 30°44′E | 10 |
| Kahramanmaraş | 37°30′N | 36°57′E | 13 | |
| Antalya | 36°50′N | 30°13′E | 30 | |
| Eastern Anatolia (EAR) | Kars | 40°36′N | 43°07′E | 30 |
| South East Anatolia (SAR) | Mardin | 37°18′N | 40°44′E | 40 |
| Şanlıurfa | 37°10′N | 38°50′E | 31 | |
| Total | 314 | |||
| Name(s) | Chromosome | Primer Sequence (5’→ 3’) | Annealing Temperature (°C) | GenBank Accession Number | Allele Range (bp) | Repeat Motif | Multiplex Group | Ref. |
|---|---|---|---|---|---|---|---|---|
| HMS07 | 1 | F:FAM-CAGGAAACTCATGTTGATACCATC R: TGTTGTTGAAACATACCTTGACTGT | 58 | X74636 | 160–178 | (AC)2(CA)n | 1 | [8] |
| ASB23 | 3 | F: NED-GAGGTTTGTAATTGGAATG R: GAGAAGTCATTTTTAACACCT | 58 | X93537 | 128–154 | (TG)17 | 1 | [9] |
| HTG07 | 4 | F: PET-CCTGAAGCAGAACATCCCTCCTTG R: ATAAAGTGTCTGGGCAGAGCTGCT | 58 | AF142607 | 272–297 | (GT)n | 3 | [10] |
| AHT05 | 8 | F: PET-ACGGACACATCCCTGCCTGC R: GCAGGCTAAGGGGGCTCAGC | 58 | - | 130–146 | (GT)n | 3 | [11] |
| HMS03 | 9 | F:NED-CCAACTCTTTGTCACATAACAAGA R: CCATCCTCACTTTTTCACTTTGTT | 58 | X74632 | 150–170 | (TG)2(CA)2TC(CA)n/(TG)2(CA)2TC(CA)Nga(CA)5 | 2 | [8] |
| HMS02 | 10 | F: NED-ACGGTGGCAACTGCCAAGGAAG R: CTTGCAGTCGAATGTGTATTAAATG | 58 | X74631 | 218–238 | (CA)n(TC)2 | 1 | [8] |
| COR058 | 12 | F: VIC-GGGAAGGACGATGAGTGAC R: CACCAGGCTAAGTAGCCAAAG | 56 | AF108375 | 210–230 | i(TG)23 | 1 | [12] |
| VHL209 | 14 | F: FAM-TCTTACATCCTTCCATTACAACTA R: TGATACATATGTACGTGAAAGGAT | 56 | Y08451 | 84–96 | (AC)17 | 2 | [13] |
| ASB02 | 15 | F: FAM-CCTTCCGTAGTTTAAGCTTCTG R: CACAACTGAGTTCTCTGATAGG | 54 | X93516 | 222–254 | (GT)24 | 2 | [14] |
| HMS20 | 16 | F: VIC-TGGGAGAGGTACCTGAAATGTAC R: GTTGCTATAAAAAATTGTCTCCCTAC | 58 | - | 116–140 | - | 2 | [15] |
| COR007 | 17 | F: PET-GTGTTGGATGAAGCGAATGA R: GACTTGCCTGGCTTTGAGTC | 56 | AF083450 | 156–170 | (GT)18 | 2 | [16] |
| LEX54 | 18 | F: FAM-TGCATGAGCCAATTCCTTAT R: TGGACAGATGACAGCAGTTC | 55 | AF075656 | 165–177 | (AC)18 | 3 | [17] |
| HTG06 | 15 | F:FAM-CCTGCTTGGAGGCTGTGATAAGAT R: GTTCACTGAATGTCAAATTCTGCT | 58 | - | 84–106 | (TG)n | 4 | [18] |
| HTG10 | 21 | F: VIC-CAATTCCCGCCCCACCCCCGGCA R: TTTTTATTCTGATCTGTCACATTT | 54 | AF169294 | 93–113 | (TG)n/TATC(TG)n | 3 | [10] |
| COR018 | 25 | F: FAM-AGTCTGGCAATATTGAGGATGT R: AGCAGCTACCCTTTGAATACTG | 56 | AF083461 | 249–271 | İ(CA)18 | 4 | [16] |
| COR071 | 26 | F: PET-CTTGGGCTACAACAGGGAATA R: CTGCTATTTCAAACACTTGGA | 56 | AF142608 | 190–202 | İ(TG)17/İ(AG)18 | 4 | [19] |
| COR082 | 29 | F: GCTTTTGTTTCTCAATCCTAGC R: TGAAGTCAAATCCCTGCTTC | 58 | AF154935 | 192–226 | (AG)n | 4 | [20] |
| Region 1 | Location 1 | N | Na | Ne | Np | HO | HE | Fıs |
|---|---|---|---|---|---|---|---|---|
| MRM | KIR | 30 | 6.706 | 3.684 | 4 | 0.6266 | 0.6893 | 0.0923 *** |
| CAT | 20 | 6.294 | 3.712 | - | 0.6882 | 0.6946 | 0.0094 | |
| MAL | 10 | 5.118 | 3.274 | 1 | 0.6379 | 0.6806 | 0.0660 | |
| BSR | MER | 30 | 6.294 | 3.618 | - | 0.6720 | 0.6851 | 0.0194 |
| TOK | 10 | 5.000 | 3.207 | - | 0.6588 | 0.6548 | −0.0065 | |
| KAS | 10 | 4.941 | 3.284 | - | 0.7118 | 0.6762 | −0.0557 | |
| AER | KUT | 10 | 4.588 | 3.215 | - | 0.6941 | 0.6755 | −0.0291 |
| MUG | 19 | 5.824 | 3.457 | 1 | 0.6563 | 0.6409 | 0.0242 | |
| AYD | 6 | 4.529 | 3.209 | 1 | 0.7139 | 0.6863 | 0.0424 | |
| CAR | KON | 15 | 5.471 | 3.399 | - | 0.6667 | 0.6830 | 0.0247 |
| MDR | ISP | 10 | 5.118 | 3.219 | - | 0.6610 | 0.6294 | 0.0503 |
| KAH | 13 | 5.529 | 3.599 | 1 | 0.6886 | 0.6670 | 0.0325 | |
| ANT | 30 | 6.471 | 3.587 | 1 | 0.6887 | 0.6983 | −0.0142 | |
| EAR | KAR | 30 | 6.471 | 3.764 | 3 | 0.6919 | 0.6819 | 0.0148 |
| SAR | MAR | 40 | 6.647 | 3.731 | 1 | 0.6888 | 0.6791 | 0.0143 |
| SAN | 31 | 6.412 | 3.803 | - | 0.6863 | 0.6831 | 0.0046 |
| Locus | TNA | Na | Ne | PIC | FIS | FST | FIT | HO | HE | HT | Hs | DST | GST | HWE | SR | F(null) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HMS07 | 7 | 3.938 | 2.988 | 0.390 | −0.0602 | 0.0201 | −0.0389 | 0.449 | 0.438 | 0.447 | 0.436 | 0.011 | 0.025 | NS | 160–178 | 0.0341 |
| ASB23 | 9 | 5.250 | 4.586 | 0.740 | 0.0169 | 0.0103 | 0.0270 | 0.737 | 0.757 | 0.763 | 0.758 | 0.005 | 0.006 | NS | 153–169 | 0.0106 |
| HMS02 | 10 | 5.813 | 4.243 | 0.960 | 0.0058 | 0.0137 | 0.0195 | 0.716 | 0.708 | 0.716 | 0.708 | 0.007 | 0.010 | NS | 221–243 | 0.0087 |
| COR058 | 11 | 6.438 | 4.921 | 0.760 | 0.0239 | 0.0128 | 0.0364 | 0.747 | 0.762 | 0.778 | 0.763 | 0.016 | 0.020 | NS | 187–209 | 0.0143 |
| HMS03 | 8 | 4.438 | 3.276 | 0.540 | 0.0538 | 0.0034 | 0.0570 | 0.579 | 0.615 | 0.623 | 0.616 | 0.007 | 0.011 | NS | 149–169 | 0.0525 |
| VHL209 | 9 | 4.313 | 3.228 | 0.450 | −0.0100 | 0.0140 | 0.0041 | 0.473 | 0.467 | 0.476 | 0.466 | 0.010 | 0.021 | NS | 76–92 | 0.0000 |
| ASB02 | 4 | 2.500 | 2.154 | 0.360 | −0.0241 | 0.0008 | −0.0232 | 0.443 | 0.445 | 0.445 | 0.446 | −0.001 | −0.002 | NS | 157–163 | 0.0000 |
| HMS20 | 7 | 5.500 | 3.794 | 0.600 | −0.0028 | 0.0000 | −0.0032 | 0.666 | 0.663 | 0.665 | 0.663 | 0.001 | 0.002 | NS | 115–131 | 0.0097 |
| COR007 | 7 | 5.375 | 4.588 | 0.710 | 0.0123 | 0.0124 | 0.0246 | 0.717 | 0.717 | 0.734 | 0.718 | 0.017 | 0.023 | NS | 165–177 | 0.0144 |
| HTG07 | 11 | 8.813 | 6.164 | 0.810 | 0.0211 | 0.0033 | 0.0244 | 0.802 | 0.827 | 0.833 | 0.828 | 0.005 | 0.006 | NS | 136–158 | 0.0601 |
| HTG10 | 9 | 7.250 | 5.321 | 0.480 | −0.0041 | 0.0078 | 0.0038 | 0.796 | 0.787 | 0.799 | 0.787 | 0.012 | 0.015 | NS | 84–104 | 0.0583 |
| COR018 | 11 | 7.188 | 5.625 | 0.800 | −0.0198 | 0.0124 | −0.0072 | 0.835 | 0.804 | 0.814 | 0.803 | 0.011 | 0.014 | NS | 252–276 | 0.0044 |
| COR071 | 8 | 6.500 | 4.997 | 0.970 | 0.0125 | 0.0096 | 0.0220 | 0.756 | 0.755 | 0.765 | 0.755 | 0.010 | 0.013 | NS | 193–207 | 0.0361 |
| COR082 | 6 | 4.563 | 3.955 | 0.650 | 0.2959 | 0.1955 | 0.4336 | 0.418 | 0.582 | 0.701 | 0.587 | 0.114 | 0.162 | NS | 214–224 | 0.1708 |
| HTG06 | 4 | 3.938 | 3.741 | 0.660 | −0.0007 | 0.0081 | 0.0073 | 0.721 | 0.711 | 0.716 | 0.711 | 0.005 | 0.007 | NS | 78–84 | 0.1894 |
| LEX54 | 9 | 5.688 | 4.234 | 0.970 | −0.0003 | 0.0000 | −0.0031 | 0.679 | 0.675 | 0.673 | 0.675 | −0.002 | −0.004 | NS | 168–192 | 0.0278 |
| AHT05 | 12 | 9.625 | 6.966 | 0.980 | 0.0167 | 0.0047 | 0.0213 | 0.857 | 0.884 | 0.890 | 0.885 | 0.004 | 0.005 | NS | 130–158 | 0.0264 |
| All loci | 142 | 5.714 | - | 0.696 | 0.0194 | 0.0192 | 0.0382 | 0.658 | 0.670 | 0.696 | 0.683 | 0.014 | 0.020 |
| Source of Variation | Variance Component (Estimate) | Variance (%) | Fixation Index | p-Value a |
|---|---|---|---|---|
| Hypothesis 1: Geographical distribution | ||||
| Among groups | 0.06303 (Va) | 1.07 | ΦIS: 0.01728 | 0.0000 *** |
| Among populations within groups | 0.05650 (Vb) | 0.96 | ΦSC: 0.00966 | 0.0254 * |
| Among individuals within populations | 0.10009 (Vc) | 1.69 | ΦCT: 0.01066 | 0.0000 *** |
| Within individuals | 5.69268 (Vd) | 96.29 | ΦIT: 0.03714 | 0.0137 * |
| Hypothesis 2: Reynold’s genetic distances distribution | ||||
| Among groups | 0.12828 (Va) | 2.16 | ΦIS: 0.01728 | 0.0000 *** |
| Among populations within groups | 0.02932 (Vb) | 0.49 | ΦSC: 0.00504 | 0.0284 * |
| Among individuals within populations | 0.10009 (Vc) | 1.68 | ΦCT: 0.02156 | 0.0000 *** |
| Within individuals | 5.69268 (Vd) | 95.67 | ΦIT: 0.04331 | 0.0000 *** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yatkın, S.; Özdil, F.; Özkan Ünal, E.; Genç, S.; Kaplan, S.; Gürcan, E.K.; Arat, S.; Soysal, M.İ. Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers. Animals 2020, 10, 1093. https://doi.org/10.3390/ani10061093
Yatkın S, Özdil F, Özkan Ünal E, Genç S, Kaplan S, Gürcan EK, Arat S, Soysal Mİ. Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers. Animals. 2020; 10(6):1093. https://doi.org/10.3390/ani10061093
Chicago/Turabian StyleYatkın, Selen, Fulya Özdil, Emel Özkan Ünal, Serdar Genç, Selçuk Kaplan, Eser Kemal Gürcan, Sezen Arat, and Mehmet İhsan Soysal. 2020. "Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers" Animals 10, no. 6: 1093. https://doi.org/10.3390/ani10061093
APA StyleYatkın, S., Özdil, F., Özkan Ünal, E., Genç, S., Kaplan, S., Gürcan, E. K., Arat, S., & Soysal, M. İ. (2020). Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers. Animals, 10(6), 1093. https://doi.org/10.3390/ani10061093







