MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens
Simple Summary
Abstract
1. Introduction
2. Materials and Method
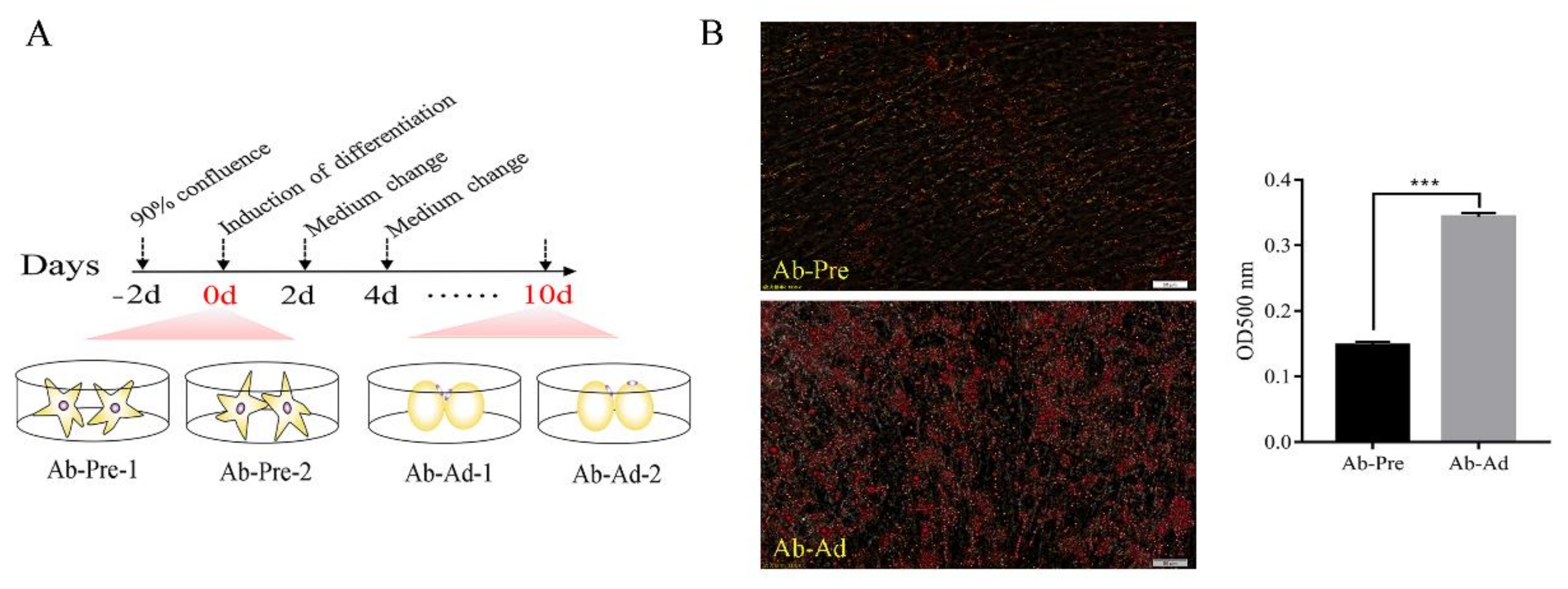
2.1. Preadipocyte Isolation, Differentiation, and Sample Collection
2.2. Oil Red O Staining
2.3. RNA Extraction, Small RNAs Library Construction, and Deep Sequencing
2.4. Data Analyses
2.5. Differential Expression Analyses
2.6. Prediction and Functional Analyses of miRNA Target Genes
2.7. Validation of miRNAs and mRNAs Expression by qRT-PCR
2.8. miRNAs–mRNAs Interaction Analysis
3. Results
3.1. Model Construction between Ab-Pre and Ab-Ad Group In Vitro
3.2. General Description of the Small RNA-seq Data
3.3. Differential Expression Analysis of miRNAs between Ab-Pre and Ab-Ad Groups
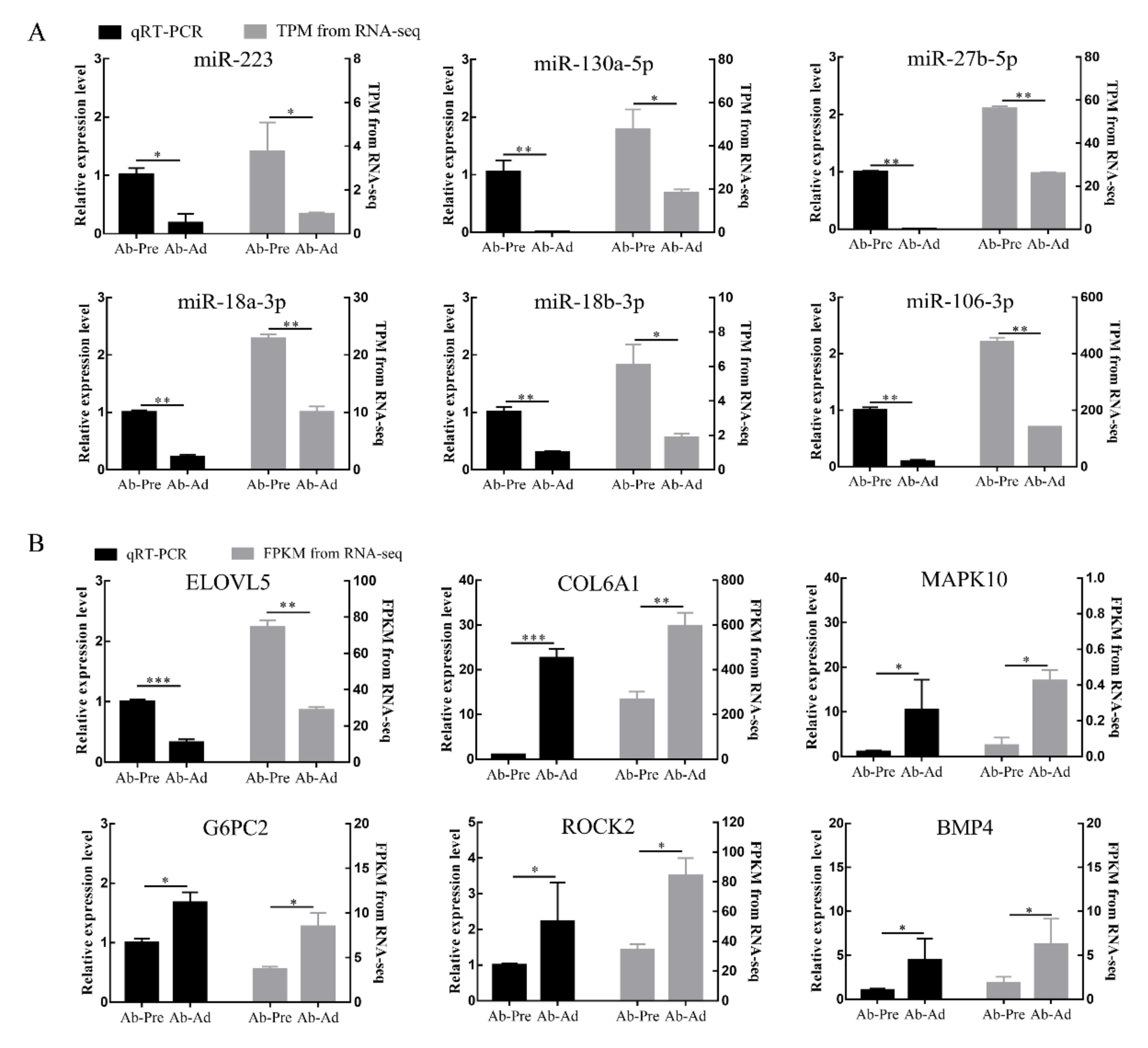
3.4. qRT-PCR Validation of the Sequencing Data
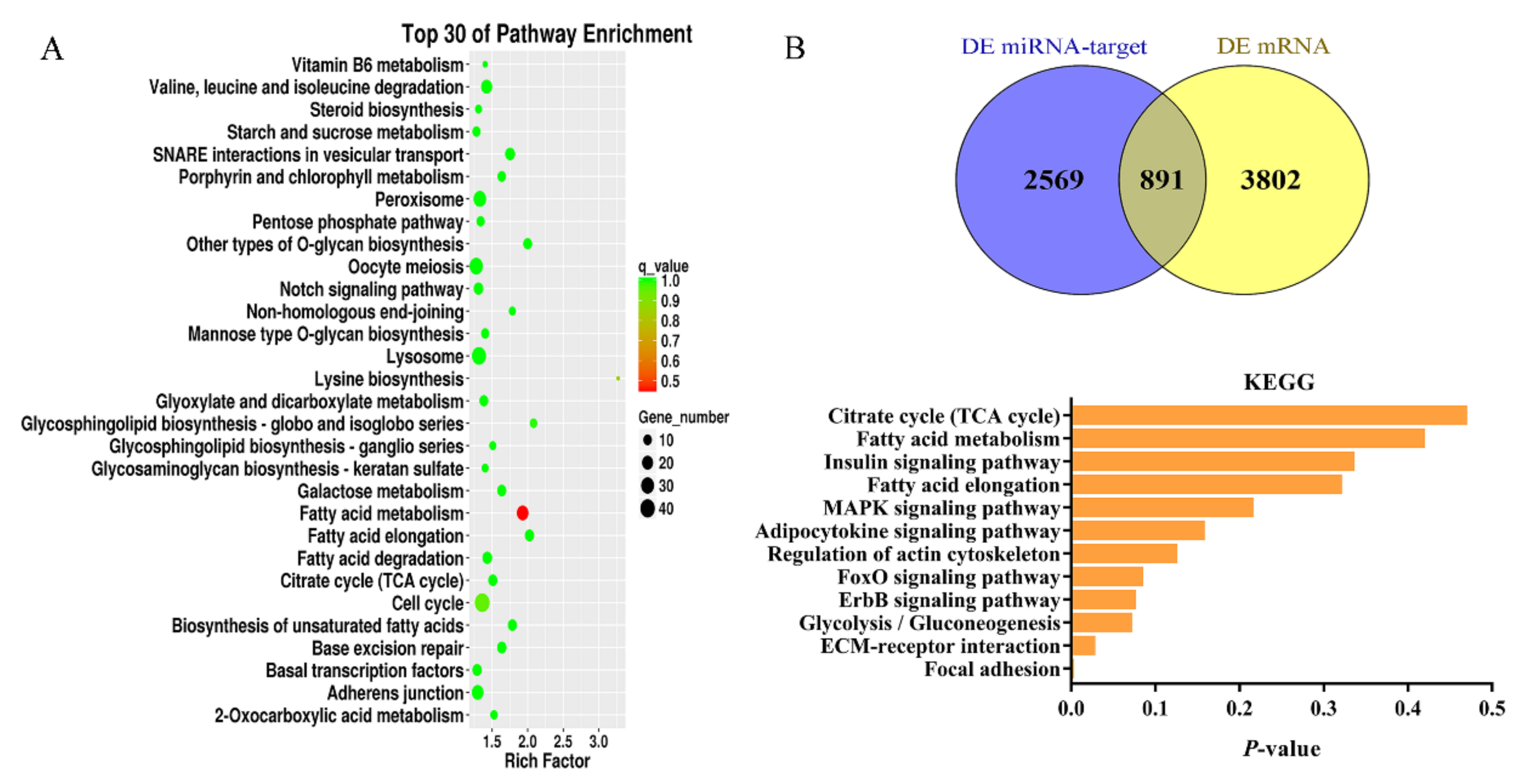
3.5. Prediction of Target Genes of Differentially Expressed miRNAs and Functional Annotation
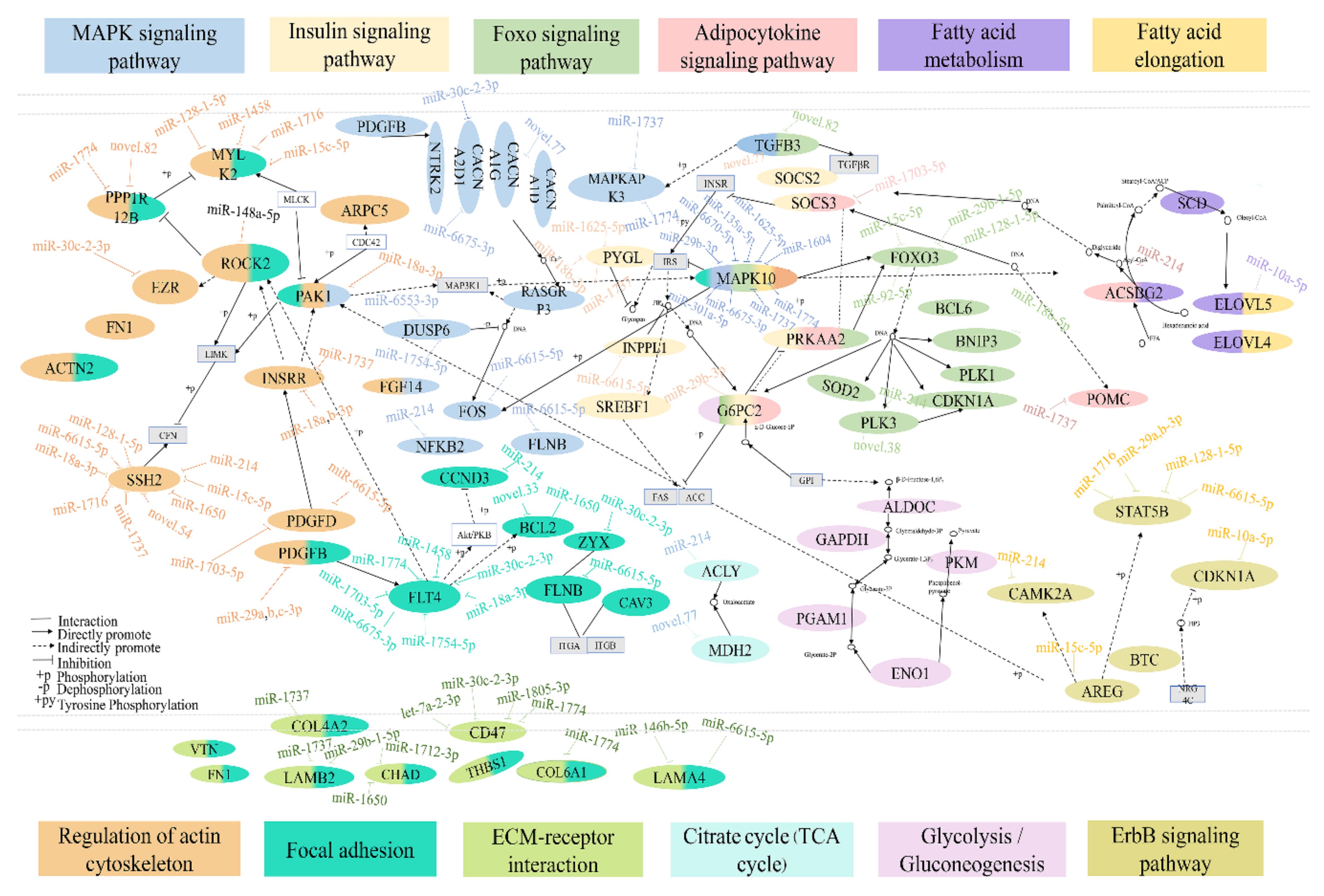
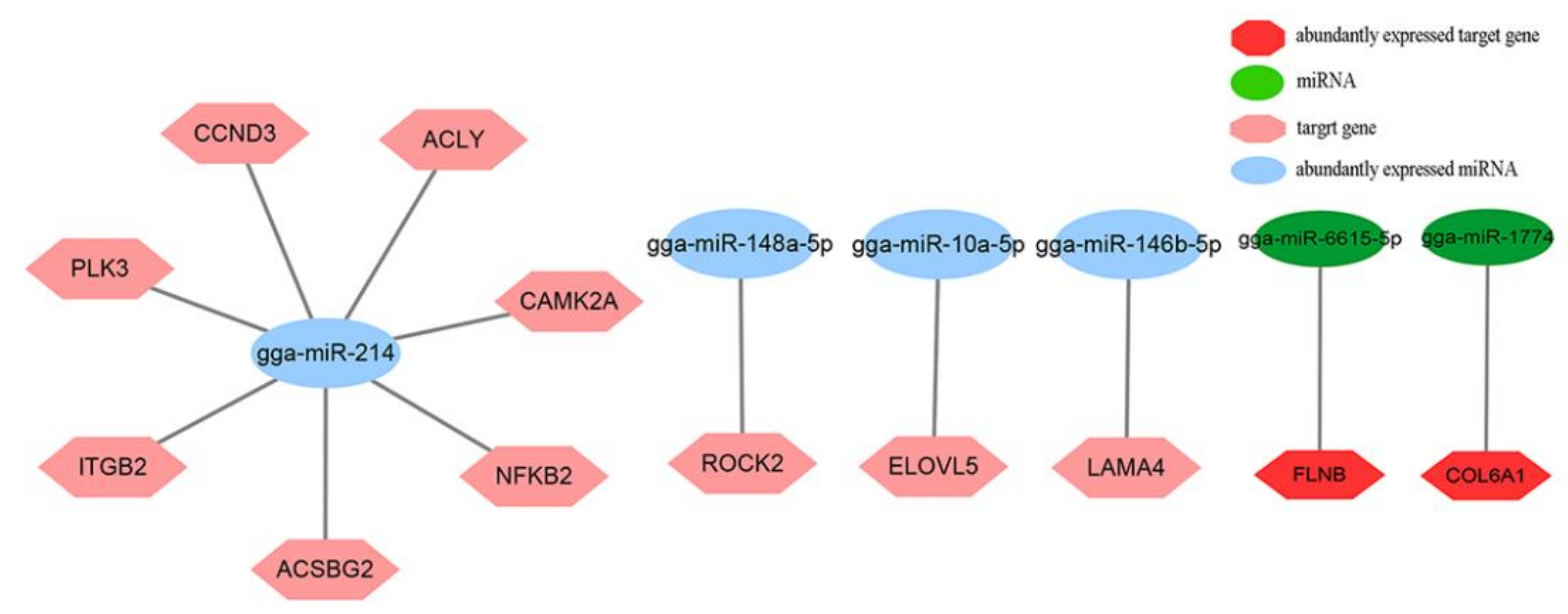
3.6. Integrated Analysis of DEMs and DEGs Involved in Lipid Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Zhang, T.; Zhang, X.; Han, K.; Zhang, G.; Wang, J.; Xie, K.; Xue, Q. Genome-Wide Analysis of lncRNA and mRNA Expression During Differentiation of Abdominal Preadipocytes in the Chicken. G3 Genesgenet. 2017, 7, 953–966. [Google Scholar] [CrossRef]
- Zhou, H.; Deeb, N.; Evock-Clover, C.M.; Ashwell, C.M.; Lamont, S.J. Genome-wide linkage analysis to identify chromosomal regions affecting phenotypic traits in the chicken. II. Body composition. Poult. Sci. 2006, 85, 1712–1721. [Google Scholar] [CrossRef]
- Kopelman, P.G. Obesity as a medical problem. Nature 2000, 404, 635–643. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. What We Talk About When We Talk About Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef]
- Zhang, M.; Li, F.; Ma, X.-F.; Li, W.-T.; Jiang, R.-R.; Han, R.-L.; Li, G.-X.; Wang, Y.-B.; Li, Z.-Y.; Tian, Y.-D. Identification of differentially expressed genes and pathways between intramuscular and abdominal fat-derived preadipocyte differentiation of chickens in vitro. BMC Genom. 2019, 20, 1–15. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Carrington, J.C.; Ambros, V. Role of MicroRNAs in Plant and Animal Development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Mullany, L.E.; Herrick, J.S.; Sakoda, L.C.; Samowitz, W.; Slattery, M.L. miRNA involvement in cell cycle regulation in colorectal cancer cases. Genes Cancer 2018, 9, 53–65. [Google Scholar]
- Saferding, V.; Hofmann, M.; Brunner, J.S.; Militaru, M.F.; Blüml, S. SAT0072 Mirna-146a is a key player in bone metabolism and osteoporosis. In Proceedings of the Annual European Congress of Rheumatology, EULAR 2018, Amsterdam, The Netherlands, 13–16 June 2018. [Google Scholar]
- He, H.; Cai, M.; Zhu, J.; Xiao, W.; Liu, B.; Shi, Y.; Yang, X.; Liang, X.; Zheng, T.; Hu, S. miR-148a-3p promotes rabbit preadipocyte differentiation by targeting PTEN. Vitro Cell. Dev. Biol. Anim. 2018, 54, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Yan, X.; Qiao, L.; Li, B.; Liu, W. MiR-330-5p negatively regulates ovine preadipocyte differentiation by targeting branched-chain aminotransferase 2. Anim. Sci. J. 2018, 89, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Fen-Fen, C.; Yan, X.; Ying, P.; Yun, G.; Jin, Q.; Gui-Yan, C.; Wei-Jun, P.; Gong-She, Y. miR-425-5p Inhibits Differentiation and Proliferation in Porcine Intramuscular Preadipocytes. Int. J. Mol. Sci. 2017, 18, 2101. [Google Scholar]
- Zhu, C.L.; Guilin, L.I.; Yang, H.C.; Yao, M.A.; Mengzhou, L.I.; Liu, H.F. MiR-26a Promoting 3t3-l1 Adipocyte Differentiation via Targeting Regulatory PTEN. J. Agric. Biotechnol. 2017, 25, 1314–1325. [Google Scholar]
- Belarbi, Y.; Mejhert, N.; Gao, H.; Arner, P.; Rydén, M.; Kulyté, A. MicroRNAs-361-5p and miR-574-5p associate with human adipose morphology and regulate EBF1 expression in white adipose tissue. Mol. Cell. Endocrinol. 2017, 472, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Cahaner, A.; Nitsan, Z. Evaluation of Simultaneous Selection for Live Body Weight and Against Abdominal Fat in Broilers. Poult. Sci. 1985, 64, 1257–1263. [Google Scholar] [CrossRef]
- Fastx. Available online: http://hannonlab.cshl.edu/fastx_toolkit/index.html (accessed on 11 March 2020).
- Toolkit Package. Available online: http://srna-workbench.cmp.uea.ac.uk/tools/analysis-tools/mircat (accessed on 11 March 2020).
- Stocks, M.B.; Moxon, S.; Mapleson, D.; Woolfenden, H.C.; Mohorianu, I.; Folkes, L.; Schwach, F.; Dalmay, T.; Moulton, V. The UEA sRNA workbench: A suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 2012, 28, 2059–2061. [Google Scholar] [CrossRef]
- Jian, Y.; Fen, Z.; Jing, L.; Jian-Ping, C.; Heng-Mu, Z.; Sek-Man, W. Integrative Analysis of the microRNAome and Transcriptome Illuminates the Response of Susceptible Rice Plants to Rice Stripe Virus. PLoS ONE 2016, 11, e0146946. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Biogeosciences 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, 149–153. [Google Scholar] [CrossRef]
- Pan, S.; Zheng, Y.; Zhao, R.; Yang, X. miRNA-374 Regulates Dexamethasone-induced Differentiation of Primary Cultures of Porcine Adipocytes. Horm. Metab. Res. 2013, 45, 518–525. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Q.; Chamba, Y.; Zhang, B.; Shang, P.; Zhang, H.; Wu, C. Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue. PLoS ONE 2017, 12, e0172930. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Lee, H.; Jung, C.H.; Jeon, T.I.; Ha, T.Y. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. Embo Mol. Med. 2013, 5, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dai, Y.-M.; Ji, C.-B.; Yang, L.; Shi, C.-M.; Xu, G.-F.; Pang, L.-X.; Huang, F.-Y.; Zhang, C.-M.; Guo, X.-R. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Mol. Cell. Endocrinol. 2014, 393, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Sun, W.; Han, H.; Chu, W.; Zhang, L.; Chen, J. miR-130a regulates differential lipid accumulation between intramuscular and subcutaneous adipose tissues of pigs via suppressing PPARG expression. Gene 2017, 636, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Lai, C.-Y.; Lin, C.-Y.; Yeh, K.-Y.; Her, G.M. MicroRNA-27b Depletion Enhances Endotrophic and Intravascular Lipid Accumulation and Induces Adipocyte Hyperplasia in Zebrafish. Int. J. Mol. Sci. 2018, 19, 93. [Google Scholar] [CrossRef]
- Li, K.; Wu, Y.; Yang, H.; Hong, P.; Fang, X.; Hu, Y. H19/miR-30a/C8orf4 axis modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. J. Cell. Physiol. 2019, 234, 20925–20934. [Google Scholar] [CrossRef]
- Wu, Y.; Zuo, J.; Zhang, Y.; Xie, Y.; Liu, F. Identification of miR-106b-93 as a Negative Regulator of Brown Adipocyte Differentiation. Biochem. Biophys. Res. Commun. 2013, 438, 575–580. [Google Scholar] [CrossRef]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef]
- Ntambi, J.M.; Young-Cheul, K. Adipocyte differentiation and gene expression. Curr. Opin. Cell Biol. 1989, 1, 1116–1121. [Google Scholar] [CrossRef]
- Niu, C.; Yuan, K.; Ma, R.; Gao, L.; Jiang, W.; Hu, X.; Lin, W.; Zhang, X.; Huang, Z. Gold nanoparticles promote osteogenic differentiation of human periodontal ligament stem cells via the p38 MAPK signaling pathway. Mol. Med. Rep. 2017, 16, 4879–4886. [Google Scholar] [CrossRef]
- Bernard, K.; Logsdon, N.J.; Benavides, G.A.; Sanders, Y.; Zhang, J.; Darley-Usmar, V.M.; Thannickal, V.J. Glutaminolysis is required for transforming growth factor-β1-induced myofibroblast differentiation and activation. J. Biol. Chem. 2018, 293, 1218–1228. [Google Scholar] [CrossRef]
- Mouw, J.K.; Guanqing, O.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Grant, R.G.; Weiss, S.J. Breaching the basement membrane: Who, when and how? Trends Cell Biol. 2008, 18, 560–574. [Google Scholar]
- Schaefer, L.; Schaefer, R.M. Proteoglycans: From structural compounds to signaling molecules. Cell Tissue Res. 2010, 339, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Sun, J.; Dong, H.; Luo, O.; Zheng, X.; Obergfell, C.; Tang, Y.; Bi, J.; O’Neill, R.; Ruan, Y. Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genom. 2014, 15, 756. [Google Scholar] [CrossRef] [PubMed]
- Mead, T.J.; Du, Y.; Nelson, C.M.; Gueye, N.A.; Drazba, J.; Dancevic, C.M.; Vankemmelbeke, M.; Buttle, D.J.; Apte, S.S. ADAMTS9-Regulated Pericellular Matrix Dynamics Governs Focal Adhesion-Dependent Smooth Muscle Differentiation. Cell Rep. 2018, 23, 485–498. [Google Scholar] [CrossRef]
- Romer, L.H.; Birukov, K.G.; Garcia, J.G.N. Focal adhesions: Paradigm for a signaling nexus. Circ. Res. 2006, 98, 606. [Google Scholar] [CrossRef]
- Beyza, V.; Giuseppe, P.; Huveyda, B. Tumor suppressor genes and ROS: Complex networks of interactions. Free Radic. Biol. Med. 2012, 52, 7–18. [Google Scholar]
- Wang, H. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar]
- Al-Sulaiti, H.; Diboun, I.; Banu, S.; Al-Emadi, M.; Amani, P.; Harvey, T.M.; Dömling, A.S.; Latiff, A.; Elrayess, M.A. Triglyceride profiling in adipose tissues from obese insulin sensitive, insulin resistant and type 2 diabetes mellitus individuals. J. Transl. Med. 2018, 16, 175. [Google Scholar] [CrossRef]
- Kadota, Y.; Toriuchi, Y.; Aki, Y.; Mizuno, Y.; Kawakami, T.; Nakaya, T.; Sato, M.; Suzuki, S. Metallothioneins regulate the adipogenic differentiation of 3T3-L1 cells via the insulin signaling pathway. PLoS ONE 2017, 12, e0176070. [Google Scholar] [CrossRef]
- Huang, H.Y.; Liu, R.R.; Zhao, G.P.; Li, Q.H.; Zheng, M.Q.; Zhang, J.J.; Li, S.F.; Liang, Z.; Wen, J. Integrated analysis of microRNA and mRNA expression profiles in abdominal adipose tissues in chickens. Sci. Rep. 2015, 5, 16132. [Google Scholar] [CrossRef] [PubMed]
- Watkins, P.A.; Dony, M.; Zhenzhen, J.; Jonathan, P. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 2007, 48, 2736–2750. [Google Scholar] [CrossRef] [PubMed]
- Moriya-Sato, A.; Hida, A.; Inagawa-Ogashiwa, M.; Wada, M.R.; Sugiyama, K.; Shimizu, J.; Yabuki, T.; Seyama, Y.; Hashimoto, N. Novel Acyl-CoA Synthetase in Adrenoleukodystrophy Target Tissues. Biochem. Biophys. Res. Commun. 2000, 279, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Zhengtong, P.; Oey, N.A.; Zuidervaart, M.M.; Zhenzhen, J.; Yuanyuan, L.; Steinberg, S.J.; Smith, K.D.; Watkins, P.A. The acyl-CoA synthetase “bubblegum” (lipidosin): Further characterization and role in neuronal fatty acid beta-oxidation. J. Biol. Chem. 2003, 278, 47070–47078. [Google Scholar]
- Xiao, C.; Hong, H.; Yu, H.; Yuan, J.; Guo, C.; Cao, H.; Li, W. MiR-340 affects gastric cancer cell proliferation, cycle, and apoptosis through regulating SOCS3/JAK-STAT signaling pathway. Immunopharmacol. Immunotoxicol. 2018, 40, 278–283. [Google Scholar] [CrossRef]
- Bjorbak, C.; Lavery, H.J.; Bates, S.H.; Olson, R.K.; Davis, S.M.; Flier, J.S.; Myers, M.G. SOCS3 mediates feedback inhibition of the leptin receptor via Tyr985. J. Biol. Chem. 2000, 275, 40649. [Google Scholar] [CrossRef]
- Marcello, P.; Mario, C.; Eliana, P.; Monica, S.; Roberta, R.; Daniela, S.; Lori, C.; Vincenzo, L.M.; Francesco, M.; Flora, M. Suppressor of cytokine signaling 3 (SOCS3) expression and hepatitis C virus-related chronic hepatitis: Insulin resistance and response to antiviral therapy. Hepatology 2010, 46, 1009–1015. [Google Scholar]
- Mori, H.; Hanada, R.; Hanada, T.; Aki, D.; Mashima, R.; Nishinakamura, H.; Torisu, T.; Chien, K.R.; Yasukawa, H.; Yoshimura, A. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat. Med. 2014, 10, 739–743. [Google Scholar] [CrossRef]
- Kumar, R.; Sanawar, R.; Li, X.; Li, F. Structure, biochemistry, and biology of PAK kinases. Gene 2017, 605, 20–31. [Google Scholar] [CrossRef]
- Sells, M.A.; Knaus, U.G.; Bagrodia, S.; Ambrose, D.M.; Bokoch, G.M.; Chernoff, J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 1997, 7, 202–210. [Google Scholar] [CrossRef]
- Delorme, V.; Machacek, M.; Dermardirossian, C.; Anderson, K.L.; Wittmann, T.; Hanein, D.; Waterman-Storer, C.; Danuser, G.; Bokoch, G.M. Cofilin Activity Downstream of Pak1 Regulates Cell Protrusion Efficiency by Organizing Lamellipodium and Lamella Actin Networks. Dev. Cell 2007, 13, 646–662. [Google Scholar] [CrossRef] [PubMed]
- Sells, M.A.; Boyd, J.T.; Chernoff, J. P21-activated kinase 1 (Pak1) regulates cell motility in mammalian fibroblasts. J. Cell Biol. 1999, 145, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Vallerie, S.N.; Hotamisligil, G.K.S. The role of JNK proteins in metabolism. Sci. Transl. Med. 2010, 2, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Yen-Yi, H.; Li, H.; Courtney, N.B.; Aravinda, C. Copy number variants in candidate genes are genetic modifiers of Hirschsprung disease. PLoS ONE 2011, 6, e21219. [Google Scholar]
- Mei, L.; Bo, L.; Huang, Y.; Yang, M.; Lan, X.; Lei, C.; Qu, W.; Bai, Y.; Chen, H. Copy number variation of bovine MAPK10 modulates the transcriptional activity and affects growth traits. Livest. Sci. 2016, 194, 44–50. [Google Scholar]
- Leah, Y.; Liu, C.S.F.; Trista, E. North, Wolfram Goessling. Functional validation of GWAS gene candidates for abnormal liver function during zebrafish liver development. Dis. Model. Mech. 2013, 6, 1271–1278. [Google Scholar]
- Hosseinibarkooie, S.; Schneider, S.; Wirth, B. Advances in understanding the role of disease-associated proteins in spinal muscular atrophy. Expert Rev. Proteom. 2017, 14, 581–592. [Google Scholar] [CrossRef]
- Jonathan, A.E. Expression Microarray Meta-Analysis Identifies Genes Associated with Ras/MAPK and Related Pathways in Progression of Muscle-Invasive Bladder Transitional Cell Carcinoma. PLoS ONE 2013, 8, e55414. [Google Scholar]
- Kalemba, K.M.; Wang, Y.; Xu, H.; Chiles, E.; Wondisford, F.E. Glycerol induces G6pc in primary mouse hepatocytes and is the preferred substrate for gluconeogenesis both in vitro and in vivo. J. Biol. Chem. 2019, 294, 18017–18028. [Google Scholar] [CrossRef]
- Hutton, J.C.; O’Brien, R.M. Glucose-6-phosphatase catalytic subunit gene family. J. Biol. Chem. 2009, 284, 29241–29245. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; Macmenamin, P.; Piedade, I.D.; Gunsalus, K.C.; Stoffel, M. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, J.; Chen, W.K.; Liu, S.; Liu, J.H.; Zhang, J.S.; Fang, K.W. MicroRNA-214 Affects Fibroblast Differentiation of Adipose-Derived Mesenchymal Stem Cells by Targeting Mitofusin-2 during Pelvic Floor Dysfunction in SD Rats with Birth Trauma. Cell. Physiol. Biochem. 2017, 42, 1870–1887. [Google Scholar] [CrossRef]
- Guo, Y.; Li, L.; Gao, J.; Chen, X.; Sang, Q. miR-214 suppresses the osteogenic differentiation of bone marrow-derived mesenchymal stem cells and these effects are mediated through the inhibition of the JNK and p38 pathways. Int. J. Mol. Med. 2017, 39, 71–80. [Google Scholar] [CrossRef]
- Du, K.T.; Deng, J.Q.; He, X.G.; Liu, Z.P.; Peng, C.; Zhang, M.S. MiR-214 Regulates the Human Hair Follicle Stem Cell Proliferation and Differentiation by Targeting EZH2 and Wnt/β-Catenin Signaling Way In Vitro. Tissue Eng. Regen. Med. 2018, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Huang, G.; Zhao, Y.; Yue, X.; Wu, H.; Li, J.; Zhu, J.; Shen, Z.; Haffty, B.G. Cullin3–KLHL25 ubiquitin ligase targets ACLY for degradation to inhibit lipid synthesis and tumor progression. Genes Dev. 2016, 30, 1956–1970. [Google Scholar] [CrossRef]
- Hien, F.N.; Kin, F.C.; Kok-Gan, C.; Ngeow, Y.F. The mRNA expression of soluble urokinase plasminogen activator surface receptor in human adipose tissue is positively correlated with body mass index. Genome 2015, 58, 315–321. [Google Scholar]
- Shi, C.; Lingxia, P.; Chenbo, J.; Jiaqin, W.; Ning, L.; Jiantao, C.; Ling, C.; Lei, Y.; Fangyan, H.; Yahui, Z. Obesity-associated miR-148a is regulated by cytokines and adipokines via a transcriptional mechanism. Mol. Med. Rep. 2016, 14, 5707–5712. [Google Scholar] [CrossRef]
- Diep, D.T.V.; Hong, K.; Khun, T.; Zheng, M.; ul-Haq, A.; Jun, H.-S.; Kim, Y.-B.; Chun, K.-H. Anti-adipogenic effects of KD025 (SLx-2119), a ROCK2-specific inhibitor, in 3T3-L1 cells. Sci. Rep. 2018, 8, 2477. [Google Scholar] [CrossRef]
- Kilian, K.A.; Bugarija, B.; Lahn, B.T.; Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 4872–4877. [Google Scholar] [CrossRef]
- Wehrle-Haller, B. Structure and function of focal adhesions. Curr. Opin. Cell Biol. 2012, 24, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Choi, P.S.; Chaffer, C.L.; Labella, K.; Hahn, W.C. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. ELife Sci. 2018, 7, e37184. [Google Scholar] [CrossRef] [PubMed]

| Samples | Raw Reads | Clean Reads | Clean Ratio (%) | Mapped Reads | Mapped Ratio (%) | Q20 Value |
|---|---|---|---|---|---|---|
| Ab-Pre-1 | 24317079 | 20332045 | 83.61% | 14782763 | 72.3% | 99.40% |
| Ab-Pre-2 | 20450354 | 19600041 | 95.84% | 16123255 | 82.3% | 99.54% |
| Ab-Ad-1 | 18449530 | 18133233 | 98.29% | 15667237 | 86.4% | 99.24% |
| Ab-Ad-2 | 20749337 | 20329584 | 97.98% | 17709466 | 87.1% | 99.59% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Sun, J.; Zhu, S.; Du, Z.; Li, D.; Li, W.; Li, Z.; Tian, Y.; Kang, X.; Sun, G. MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens. Animals 2020, 10, 468. https://doi.org/10.3390/ani10030468
Ma X, Sun J, Zhu S, Du Z, Li D, Li W, Li Z, Tian Y, Kang X, Sun G. MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens. Animals. 2020; 10(3):468. https://doi.org/10.3390/ani10030468
Chicago/Turabian StyleMa, Xiangfei, Junwei Sun, Shuaipeng Zhu, Zhenwei Du, Donghua Li, Wenting Li, Zhuanjian Li, Yadong Tian, Xiangtao Kang, and Guirong Sun. 2020. "MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens" Animals 10, no. 3: 468. https://doi.org/10.3390/ani10030468
APA StyleMa, X., Sun, J., Zhu, S., Du, Z., Li, D., Li, W., Li, Z., Tian, Y., Kang, X., & Sun, G. (2020). MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens. Animals, 10(3), 468. https://doi.org/10.3390/ani10030468





