Association Analysis to Copy Number Variation (CNV) of Opn4 Gene with Growth Traits of Goats
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Sample Collection and DNA Extraction
2.3. Design and Detection of Primers
2.4. Statistical Analysis
3. Results
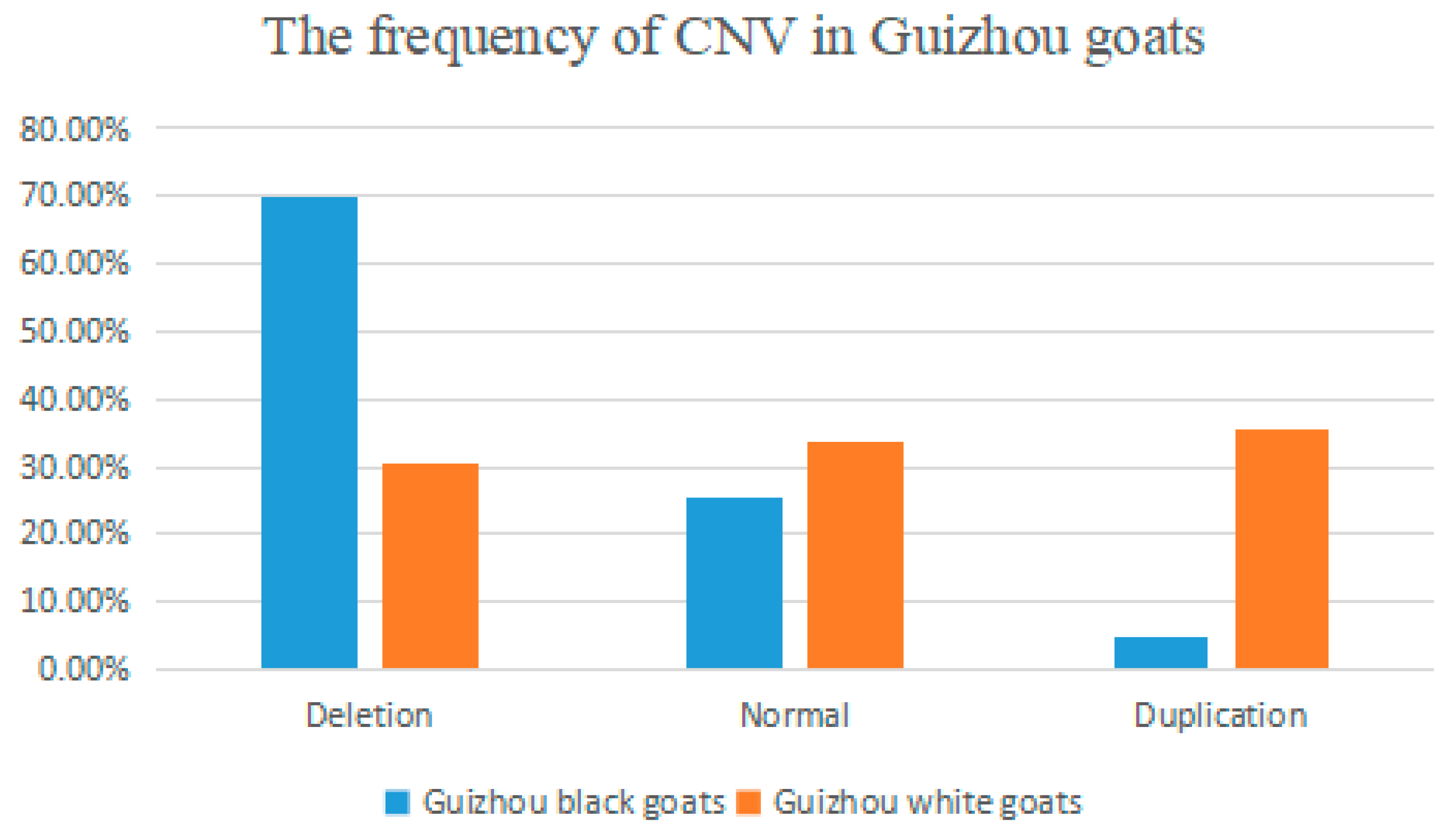
3.1. Distribution of Different CNV Types in Goats
3.2. Correlation Analysis of CNV Type and Goat Growth Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, T.M.; Tsujimura, S.; Allen, A.E.; Wynne, J.; Bedford, R.; Vickery, G.; Vugler, A.; Lucas, R.J. Melanopsin-based brightness discrimination in mice and humans. Curr. Biol. 2012, 22, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Provencio, I.; Jiang, G.; De Grip, W.J.; Hayes, W.P.; Rollag, M.D. Melanopsin: An opsin in melanophores, brain, and eye. Proc. Natl. Acad. Sci. USA 1998, 95, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.W.; Hannibal, J.; Hagiwara, G.; Colas, D.; Ruppert, E.; Ruby, N.F.; Heller, H.C.; Franken, P.; Bourgin, P. Melanopsin as a sleep modulator: Circadian gating of the direct effects of light on sleep and altered sleep homeostasis in Opn4(−/−) mice. PLoS Biol. 2009, 7, e1000125. [Google Scholar] [CrossRef] [PubMed]
- Ondrusova, K.; Fatehi, M.; Barr, A.; Czarnecka, Z. Subcutaneous white adipocytes express a light sensitive signaling pathway mediated via a melanopsin/TRPC channel axis. Sci. Rep. 2017, 7, 16332. [Google Scholar] [CrossRef] [PubMed]
- Bailes, H.J.; Lucas, R.J. Human melanopsin forms a pigment maximally sensitive to blue light (λ max ≈ 479 nm) supporting activation of Gq/11 and Gi/o signalling cascades. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122987. [Google Scholar] [CrossRef]
- Hankins, M.W.; Peirson, S.N.; Foster, R.G. Melanopsin: An exciting photopigment. Trends Neurosci. 2008, 31, 27–36. [Google Scholar] [CrossRef]
- Poteser, M.; Graziani, A.; Eder, P.; Yates, A.; Mächler, H.; Romanin, C.; Groschner, K. Identification of a rare subset of adipose tissue-resident progenitor cells, which express CD133 and TRPC3 as a VEGF-regulated Ca2+ entry channel. FEBS Lett. 2008, 582, 2696–2702. [Google Scholar] [CrossRef]
- Liu, G.E.; Hou, Y.; Zhu, B.; Cardone, M.F.; Jiang, L.; Cellamare, A.; Mitra, A.; Alexander, L.J.; Coutinho, L.L.; Dell’Aquila, M.E. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010, 20, 693–703. [Google Scholar] [CrossRef]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.L.; Chen, C.; Zhai, Y.; et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998, 20, 207. [Google Scholar] [CrossRef]
- Winchester, L.; Yau, C.; Ragoussis, J. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genom. Proteom. 2009, 8, 353–366. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, K.; Shi, X.; Li, F.; Song, L.; Wu, H.; Dean, M.; Li, G.; Tsang, S.; Jiang, R.; et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience 2015, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.; Dube, S.; Mir, A.; Qin, J.; Sun, G.; Ramakrishnan, R.; Jones, R.C.; Livak, K.J. Taking qPCR to a higher level: Analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 2010, 50, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef]
- Mefford, H.C.; Cooper, G.M.; Zerr, T.; Smith, J.D.; Baker, C.; Shafer, N.; Thorland, E.C.; Skinner, C.; Schwartz, C.E.; Nickerson, D.A.; et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009, 19, 1579–1585. [Google Scholar] [CrossRef]
- McCarroll, S.A.; Kuruvilla, F.G.; Korn, J.M.; Cawley, S.; Nemesh, J.; Wysoker, A.; Shapero, M.H.; de Bakker, P.I.; Maller, J.B.; Kirby, A.; et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008, 40, 1166. [Google Scholar] [CrossRef]
- Bae, J.S.; Cheong, H.S.; Kim, J.O.; Lee, S.O.; Kim, E.M.; Lee, H.W.; Kim, S.; Kim, J.W.; Cui, T.; Inoue, I.; et al. Identification of SNP markers for common CNV regions and association analysis of risk of subarachnoid aneurysmal hemorrhage in Japanese population. Biochem. Biophys. Res. Commun. 2008, 373, 593–596. [Google Scholar] [CrossRef]
- Porter, M.L.; Blasic, J.R.; Bok, M.J.; Cameron, E.G.; Pringle, T.; Cronin, T.W.; Robinson, P.R. Shedding new light on opsin evolution. Proc. Biol. Sci. 2012, 279, 3–14. [Google Scholar] [CrossRef]
- Díaz, N.M.; Morera, L.P.; Guido, M.E. Melanopsin and the Non-visual Photochemistry in the Inner Retina of Vertebrates. Photochem. Photobiol. 2016, 92, 29–44. [Google Scholar] [CrossRef]
- Zhou, Y.; Utsunomiya, Y.T.; Xu, L.; Hay el, H.A.; Bickhart, D.M.; Alexandre, P.A.; Rosen, B.D.; Schroeder, S.G.; Carvalheiro, R.; de Rezende Neves, H.H.; et al. Genome-wide CNV analysis reveals variants associated with growth traits in Bos indicus. BMC Genom. 2016, 17, 419. [Google Scholar] [CrossRef]
- Xu, L.; Cole, J.B.; Bickhart, D.M.; Hou, Y.; Song, J.; VanRaden, P.M.; Sonstegard, T.S.; Van Tassell, C.P.; Liu, G.E. Genome wide CNV analysis reveals additional variants associated with milk production traits in Holsteins. BMC Genom. 2014, 15, 683. [Google Scholar] [CrossRef] [PubMed]
- Ionita-Laza, I.; Rogers, A.J.; Lange, C.; Raby, B.A.; Lee, C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics 2009, 93, 22–26. [Google Scholar] [CrossRef] [PubMed]
| The Growth Traits | The Measured Method |
|---|---|
| Heart girth | The circumference of the chest behind the rear edge of scapula. |
| Withers height | Vertical distance from the highest position of shoulder to the ground. |
| Body length | The straight distance from shoulder to rear edge of ischial tuberosity. |
| Body weight | Weighing after stop feeding for a day. |
| Primer | Position | Sequence (5’->3’) | Product Length |
|---|---|---|---|
| Opn4-CNV | Forward primer | CGTGATACCAGGCTCCAGA | 19 nt |
| Reverse primer | ACGGCGAGGTTGATAATGA | ||
| MC1R | Forward primer | CTCGTTGGCCTCTTCATAGC | 21 nt |
| Reverse primer | GAAGTTCTTGAAGATGCAGCC |
| Growth Traits (Average Value ± Standard Error) | CNV Types | p Value | ||
|---|---|---|---|---|
| Deletion (n = 130) | Normal (n = 47) | Duplication (n = 9) | ||
| BW (kg) | 28.113 ± 0.749 | 28.034 ± 1.245 | 27.167 ± 2.845 | 0.95 |
| WH (cm) | 59.254 ± 0.538 | 59.426 ± 0.895 | 60.000 ± 2.045 | 0.933 |
| BL (cm) | 60.400 ± 0.588 a | 62.936 ± 0.978 b | 65.222 ± 2.235 b | 0.018 * |
| HG (cm) | 74.377 ± 0.608 | 74.170 ± 1.102 | 72.222 ± 2.312 | 0.666 |
| Growth Traits (Average Value ± Standard Error) | CNV Types | p Value | ||
|---|---|---|---|---|
| Deletion (n = 30) | Normal (n = 33) | Duplication (n = 35) | ||
| BW (kg) | 27.133 ± 1.331 a | 31.502 ± 1.269 b | 26.230 ± 1.232 a | 0.010 * |
| HG (cm) | 71.167 ± 1.214 | 73.773 ± 1.158 | 70.614 ± 1.124 | 0.123 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Yang, P.; Shi, S.; Zhang, Z.; Shi, Q.; Xu, J.; He, H.; Lei, C.; Wang, E.; Chen, H.; et al. Association Analysis to Copy Number Variation (CNV) of Opn4 Gene with Growth Traits of Goats. Animals 2020, 10, 441. https://doi.org/10.3390/ani10030441
Li L, Yang P, Shi S, Zhang Z, Shi Q, Xu J, He H, Lei C, Wang E, Chen H, et al. Association Analysis to Copy Number Variation (CNV) of Opn4 Gene with Growth Traits of Goats. Animals. 2020; 10(3):441. https://doi.org/10.3390/ani10030441
Chicago/Turabian StyleLi, LiJuan, Peng Yang, ShuYue Shi, ZiJing Zhang, QiaoTing Shi, JiaWei Xu, Hua He, ChuZhao Lei, ErYao Wang, Hong Chen, and et al. 2020. "Association Analysis to Copy Number Variation (CNV) of Opn4 Gene with Growth Traits of Goats" Animals 10, no. 3: 441. https://doi.org/10.3390/ani10030441
APA StyleLi, L., Yang, P., Shi, S., Zhang, Z., Shi, Q., Xu, J., He, H., Lei, C., Wang, E., Chen, H., & Huang, Y. (2020). Association Analysis to Copy Number Variation (CNV) of Opn4 Gene with Growth Traits of Goats. Animals, 10(3), 441. https://doi.org/10.3390/ani10030441





