Genome-Wide Association Study Reveals Candidate Genes for Litter Size Traits in Pelibuey Sheep
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals Used and Obtaining Samples
2.2. Extraction of DNA and Genotyping
2.3. Genotyping Analysis and Data Quality Control
2.4. Genome-Wide Association Analysis
2.5. Candidate Gene and QTLs Associated
2.6. Analysis of Gene Ontology and Metabolic Pathways
3. Results
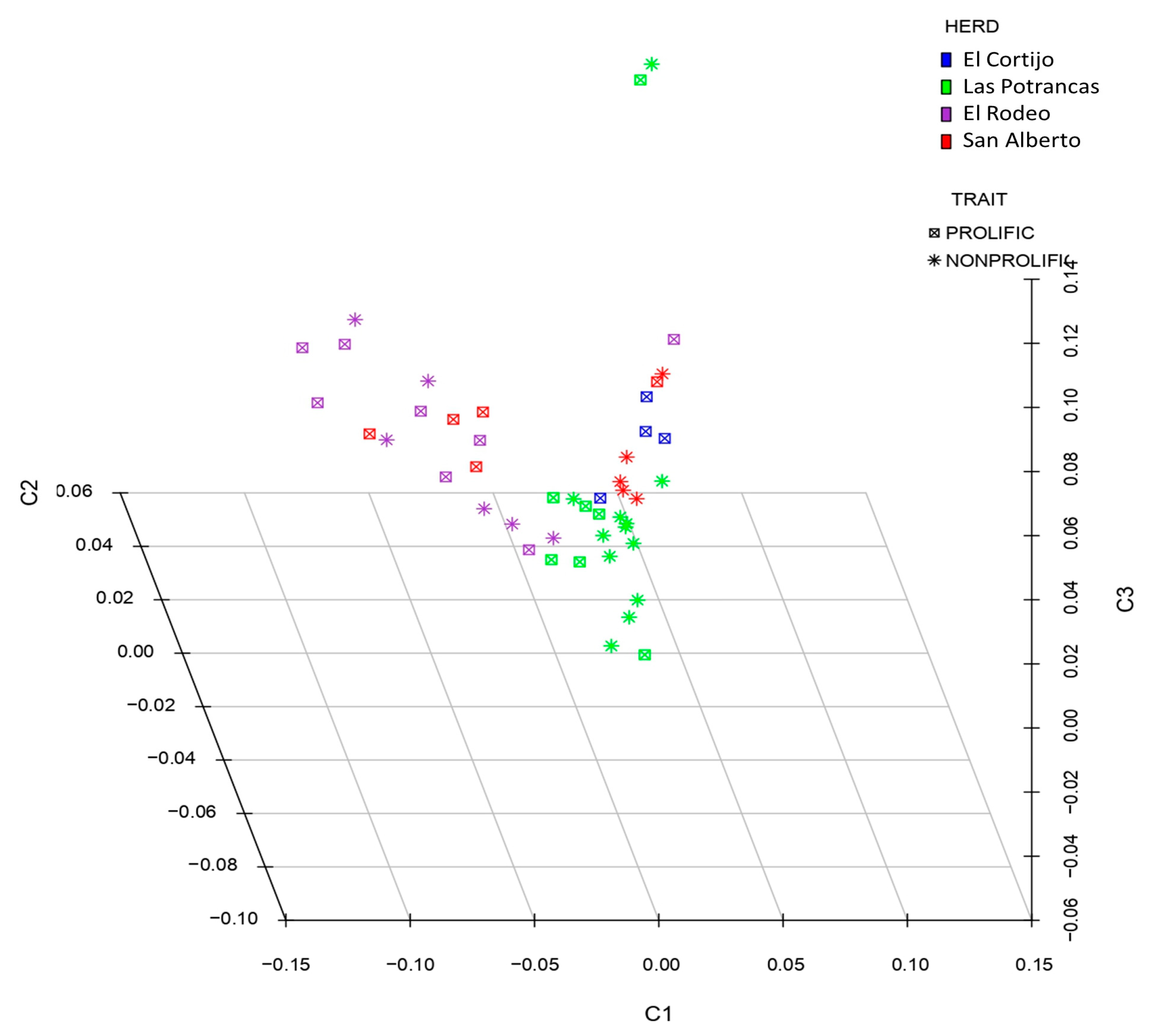
3.1. Genotyping Analysis and Data Quality Control
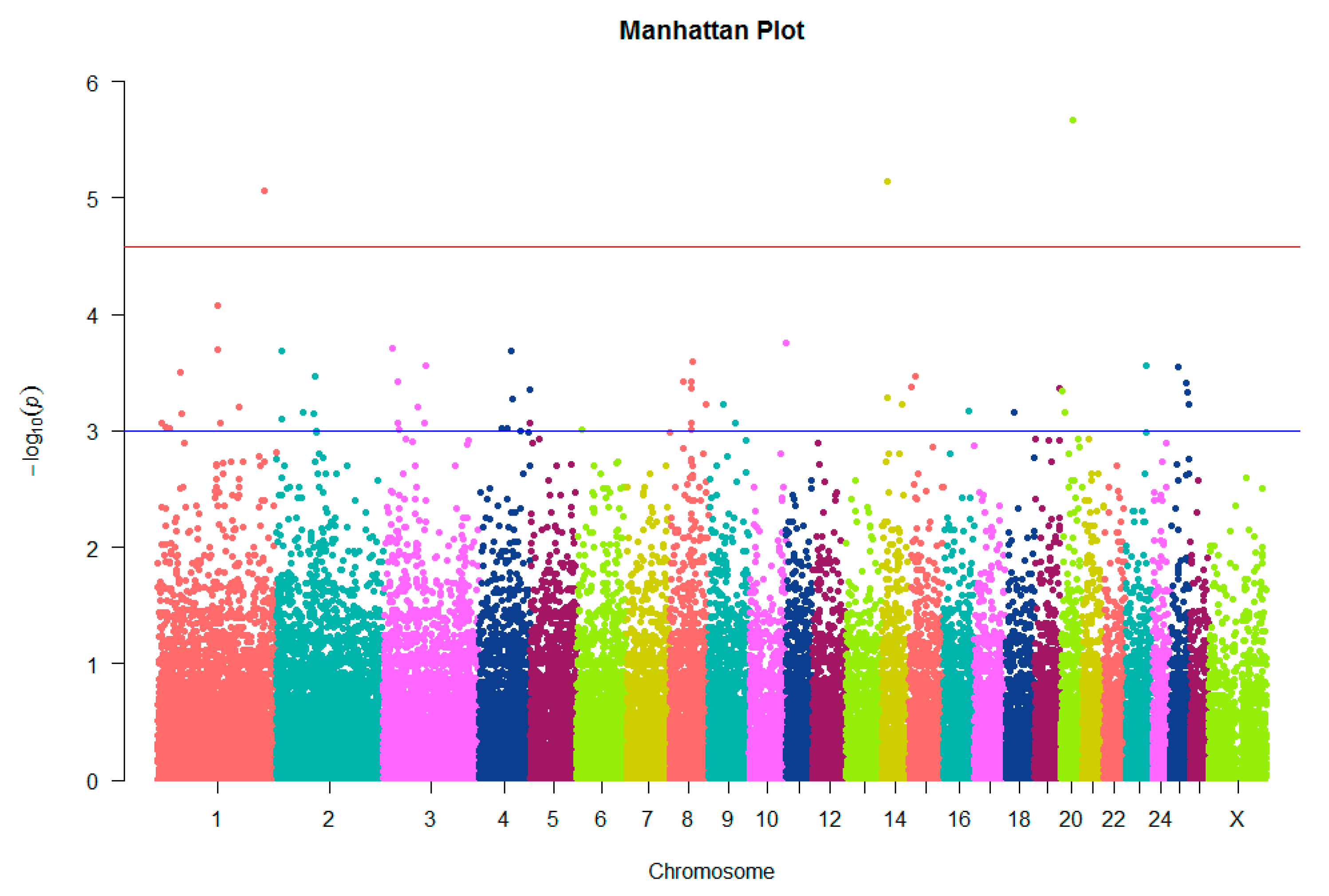
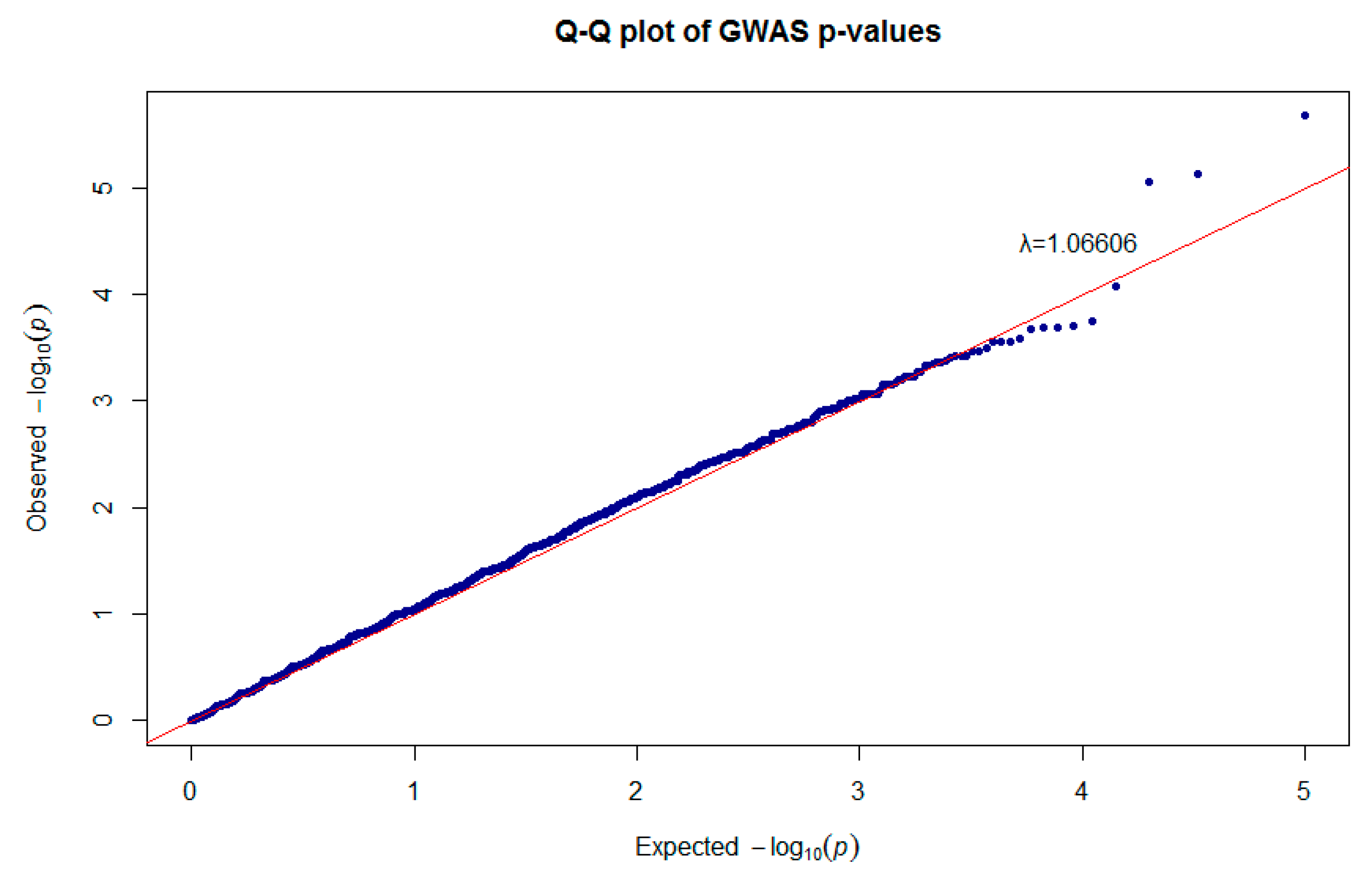
3.2. Genome-Wide Association Analysis
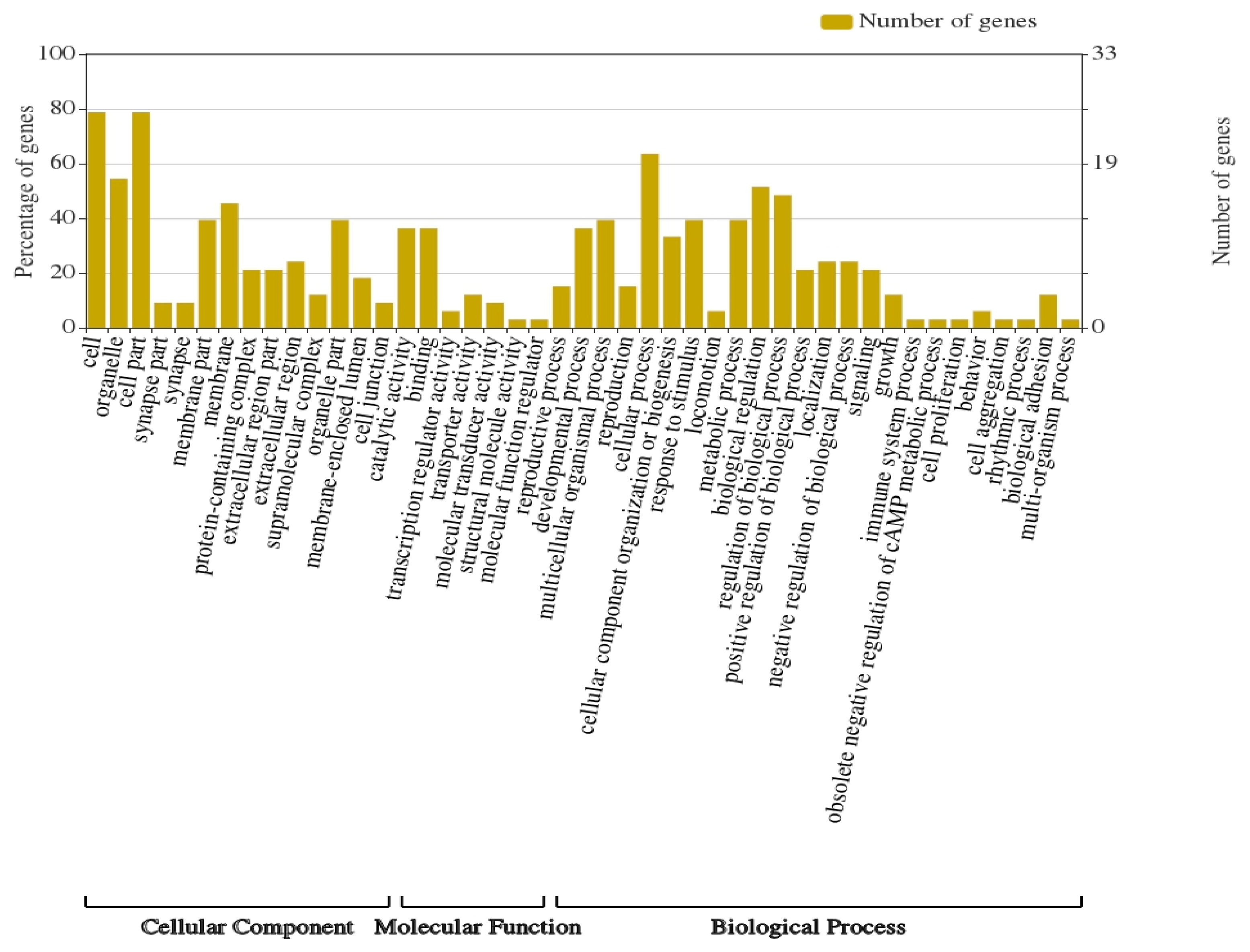
3.3. Gene Ontology and KEGG Analysis
3.4. Identification of the Candidate Genes
4. Discussion
4.1. Markers Associated with Prolificacy Traits
4.2. Gene Ontology and Metabolic Pathways
4.3. Candidate Gene Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bobadilla-Soto, E.E.; Salas-Razo, G.; Padillas-Flores, J.P.; Perea-Peña, M. Unit displacement of sheep production in Mexico by effect of imports. Int. J. Dev. Res. 2018, 5, 3607–3612. [Google Scholar]
- Macías-Cruz, U.; Álvarez-Valenzuela, F.D.; Correa-Calderon, A.; Molina-Ramirez, L.; González-Reyna, A.; Soto-Navarro, S.; Avendaño-Reyes, L. Pelibuey ewe productivity and subsequent pre-weaning lamb performance using hair-sheep breeds under a confinement system. J. Appl. Anim. Res. 2009, 36, 255–260. [Google Scholar] [CrossRef]
- Macías-Cruz, U.; Álvarez-Valenzuela, F.; Olguín-Arredondo, H.; Molina-Ramirez, L.; Avendaño-Reyes, L. Ovejas Pelibuey sincronizadas con progestágenos y apareadas con machos de razas Dorper y Katahdin bajo condiciones estabuladas: Producción de la oveja y crecimiento de los corderos durante el período predestete. Arch. Med. Vet. 2012, 44, 29–37. [Google Scholar] [CrossRef]
- Dickson, L.; Torres, G.; Aubeterre, R.D.; Garcia, O. Factores que influyen en el intervalo entre partos y la prolificidad de un hato de carneros Pelibuey en Venezuela. Rev. Cuba. Cienc. Agric. 2004, 38, 13–17. [Google Scholar]
- Aké-López, R.; Segura-Correa, J.C.; Quintal-Franco, J. Effect of flunixin meglumine on the corpus luteum and possible prevention of embryonic loss in Pelibuey ewes. Small Rumin. Res. 2005, 59, 83–87. [Google Scholar] [CrossRef]
- Davis, G.H.; Balakrishnan, L.; Ross, I.K.; Wilson, T.; Galloway, S.M.; Lumsden, B.M.; Hanrahan, J.P.; Mullen, M.; Mao, X.Z.; Wang, G.L.; et al. Investigation of the Booroola (FecB) and Inverdale (FecX I) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim. Reprod. Sci. 2006, 92, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo, L.V.; Ponz, R.; Tejedor, M.T.; Laviña, A.; Sierra, I. A 17 bp deletion in the Bone Morphogenetic Protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed. Anim. Reprod. Sci. 2009, 110, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.J.H.; MacDougall, C.; Campbell, B.K.; McNeilly, A.S.; Baird, D.T. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. Rapid Comun. 2001, 169, R1–R6. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Royo, A.; Jurado, J.J.; Smulders, J.P.; Marti, J.I.; Alabart, J.L.; Roche, A.; Fantova, E.; Bodin, L.; Mulsant, P.; Serrano, M.; et al. A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Anim. Genet. 2008, 39, 294–297. [Google Scholar] [CrossRef]
- Hanrahan, J.P.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, H.G.; Powell, R.; Galloway, M.S. Mutations in the Genes for Oocyte-Derived Growth Factors GDF9 and BMP15 Are Associated with Both Increased Ovulation Rate and Sterility in Cambridge and Belclare Sheep (Ovis aries). Biol. Reprod. 2004, 70, 900–909. [Google Scholar] [CrossRef]
- Galloway, S.M.; McNatty, K.P.; Cambridge, L.M.; Laitinen, M.P.; Juengel, J.L.; Jokiranta, T.S.; McLaren, R.J.; Luiro, K.; Dodds, K.G.; Montgomery, G.W.; et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat. Genet. 2000, 25, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Guéripel, X.; Véronique, B.; Gougeon, A. Oocyte Bone Morphogenetic Protein 15, but not Growth Differentiation Factor 9, Is Increased During Gonadotropin-Induced Follicular Development in the Immature Mouse and Is Associated with Cumulus Oophorus Expansion. Biol. Reprod. 2006, 75, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, A.; Cabau, C.; Bouchez, O.; Sarry, J.; Marsaud, N.; Foissac, S.; Woloszyn, F.; Mulsant, P.; Mandon-pepin, B. An overview of gene expression dynamics during early ovarian folliculogenesis: Specificity of follicular compartments and bi-directional dialog. BMC Genom. 2013, 14, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodin, L.; Di Pasquale, E.; Febre, S.; Bontoux, M.; Monget, P.; Persani, L.; Mulsant, P. A Novel Mutation in the Bone Morphogenetic Protein 15 Gene Causing Defective Protein Secretion Is Associated Lacaune Sheep. Endocrinology 2007, 148, 393–400. [Google Scholar] [CrossRef]
- Feary, E.S.; Juengel, J.L.; Smith, P.; French, M.C.; Connell, A.R.O.; Lawrence, S.B.; Galloway, S.M.; Davis, G.H.; Mcnatty, K.P. Patterns of Expression of Messenger RNAs Encoding GDF9, BMP15, TGFBR1, BMPR1B, and BMPR2 During Follicular Development and Characterization of Ovarian Follicular Populations in Ewes Carrying the Woodlands FecX2 W Mutation 1. Biol. Reprod. 2007, 77, 990–998. [Google Scholar] [CrossRef]
- Demars, J.; Fabre, S.; Sarry, J.; Rossetti, R.; Gilbert, H.; Persani, L.; Tosser-Klopp, G.; Mulsant, P.; Nowak, Z.; Drobik, W.; et al. Genome-Wide Association Studies Identify Two Novel BMP15 Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep. PLoS Genet. 2013, 9, 1–13. [Google Scholar] [CrossRef]
- Lassoued, N.; Benkhlil, Z.; Woloszyn, F.; Rejeb, A.; Aouina, M.; Rekik, M.; Fabre, S.; Bedhiaf-Romdhani, S. FecXBar a Novel BMP15 mutation responsible for prolificacy and female sterility in Tunisian Barbarine Sheep. BMC Genet. 2017, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Nicol, L.; Bishop, S.C.; Pong-wong, R.; Bendixen, C.; Holm, L.; Rhind, S.M.; Mcneilly, A.S. Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thaka sheep. Reprod. Res. 2009, 138, 921–933. [Google Scholar] [CrossRef] [Green Version]
- Silva, B.D.M.; Castro, E.A.; Souza, C.J.H.; Paiva, S.R.; Sartori, R.; Franco, M.M.; Azevedo, H.C.; Silva, T.A.S.N.; Vieira, A.M.C.; Neves, J.P.; et al. A new polymorphism in the Growth and Differentiation Factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim. Genet. 2010, 42, 89–92. [Google Scholar] [CrossRef] [Green Version]
- Våge, D.I.; Husdal, M.; Kent, M.P.; Klemetsdal, G.; Boman, I.A. A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep. BMC Genet. 2013, 14, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sagar, N.G.; Kumar, S.; Baranwal, A.; Prasad, A.R. Introgression of Fecundity Gene (FecB) in Non-Prolific SHEEP Breeds: A Boon for Farmers. Int. J. Sci. Environ. Technol. 2017, 6, 375–380. [Google Scholar]
- Duncan, E.L.; Brown, M.A. Genome-wide Association Studies. In Genetics of Bone Biology and Skeletal Disease; Rajesh, V., Thakker Michael, P., Whyte John, A., Eisman, T.I., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 93–100. [Google Scholar]
- Sallé, G.; Jacquiet, P.; Gruner, L.; Cortet, J.; Sauvé, C.; Prévot, F.; Grisez, C.; Bergeaud, J.P.; Schibler, L.; Tircazes, A.; et al. A genome scan for QTL affecting resistance to Haemonchus contortus in sheep. J. Anim. Sci. 2012, 4690–4705. [Google Scholar] [CrossRef] [PubMed]
- José, J.; Gouveia, D.S.; Rezende, S.; Mcmanus, C.M.; Caetano, R.; Kijas, J.W.; Facó, O.; Costa, H.; De Araujo, M.; José, C.; et al. Genome-wide search for signatures of selection in three major Brazilian locally adapted sheep breeds. Livest. Sci. 2017, 197, 36–45. [Google Scholar]
- Xu, S.-S.; Gao, L.; Xie, X.-L.; Ren, Y.-L.; Shen, Z.-Q.; Wang, F.; Shen, M.; Eyϸórsdóttir, E.; Hallsson, J.H.; Kiseleva, T.; et al. Genome-Wide Association Analyses Highlight the Potential for Different Genetic Mechanisms for Litter Size Among Sheep Breeds. Front. Genet. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Al-mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 2015, 47, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benavides, M.V.; Souza, C.J.H.; Moraes, J.C.F. How efficiently Genome-Wide Association Studies (GWAS) identify prolificity-determining genes in sheep. Genet. Mol. Res. 2018, 17, 9–14. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Wigginton, J.E.; Cutler, D.J.; Abecasis, G.R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005, 76, 887–893. [Google Scholar] [CrossRef] [Green Version]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv 2014. [Google Scholar] [CrossRef]
- Brinster, R.; Köttgen, A.; Tayo, B.O.; Schumacher, M.; Sekula, P. Control procedures and estimators of the false discovery rate and their application in low-dimensional settings: An empirical investigation. BMC Bioinform. 2018, 19, 1–10. [Google Scholar] [CrossRef]
- Nakagawa, S. A farewell to Bonferroni: The problems of low statistical power and publication bias. Behav. Ecol. 2004, 15, 1044–1045. [Google Scholar] [CrossRef]
- Balding, D.J. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006, 7, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Sahana, G.; Guldbrandtsen, B.; Bendixen, C.; Lund, M.S. Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle. Anim. Genet. 2010, 41, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavanagh, C.R.; Jonas, E.; Hobbs, M.; Thomson, P.C.; Tammen, I.; Raadsma, H.W. Mapping Quantitative Trait Loci (QTL) in sheep. III. QTL for carcass composition traits derived from CT scans and aligned with a meta-assembly for sheep and cattle carcass QTL. Genet. Sel. Evol. 2010, 42, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateescu, R.G.; Thonney, M.L. Genetic mapping of quantitative trait loci for aseasonal reproduction in sheep. Anim. Genet. 2010, 41, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gámez, E.; Gutiérrez-Gil, B.; Suarez-Vega, A.; de la Fuente, L.F.; Arranz, J.J. Identification of quantitative trait loci underlying milk traits in Spanish dairy sheep using linkage plus combined linkage disequilibrium and linkage analysis approaches. J. Dairy Sci. 2013, 96, 6059–6069. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Gil, B.; El-Zarei, M.F.; Alvarez, L.; Bayón, Y.; De La Fuente, L.F.; San Primitivo, F.; Arranz, J.J. Quantitative trait loci underlying milk production traits in sheep. Anim. Genet. 2009, 40, 423–434. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Aoki-kinoshita, K.F.; Kanehisa, M. Gene Annotation and Pathway Mapping in KEGG. Methods Mol Biol. 2007, 396, 71–91. [Google Scholar]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, w293–w297. [Google Scholar] [CrossRef] [PubMed]
- Lantier, I.; Moreno, C.R.; Berthon, P.; Sallé, G.; Pitel, F.; Schibler, L.; Gautier-Bouchardon, A.V.; Boivin, R.; Weisbecker, J.L.; François, D.; et al. Quantitative trait loci for resistance to infection in sheep using a live Salmonella Abortusovis vaccine. Anim. Genet. 2012, 43, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Beh, K.J.; Hulme, D.J.; Callaghan, M.J.; Leish, Z.; Lenane, I.; Windon, R.G.; Maddox, J.F. A genome scan for quantitative trait loci affecting resistance to Trichostrongylus colubriformis in sheep. Anim. Genet. 2002, 33, 97–106. [Google Scholar] [CrossRef]
- Roldan, D.L.; Dodero, A.M.; Bidinost, F.; Taddeo, H.R.; Allain, D.; Poli, M.A.; Elsen, J.M. Merino sheep: A further look at quantitative trait loci for wool production. Animal 2010, 4, 1330–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonas, E.; Thomson, P.C.; Hall, E.J.S.; McGill, D.; Lam, M.K.; Raadsma, H.W. Mapping quantitative trait loci (QTL) in sheep. IV. Analysis of lactation persistency and extended lactation traits in sheep. Genet. Sel. Evol. 2011, 43, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, A.M.; Paterson, K.A.; Dodds, K.G.; Tascon, C.D.; Williamson, P.A.; Thomson, M.R.; Bisset, S.A.; Beattie, A.E.; Greer, G.J.; Green, R.S.; et al. Discovery of quantitative trait loci for resistance to parasitic nematode infection in sheep: I. Analysis of outcross pedigrees. BMC Genom. 2006, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamichou, E.; Richardson, R.I.; Nute, G.R.; Gibson, K.P.; Bishop, S.C. Genetic analyses and quantitative trait loci detection, using a partial genome scan, for intramuscular fatty acid composition in Scottish Blackface sheep. J. Anim. Sci. 2006, 84, 3228–3238. [Google Scholar] [CrossRef] [Green Version]
- Ponz, R.; Moreno, C.; Allain, D.; Elsen, J.M.; Lantier, F.; Lantier, I.; Brunel, J.C.; Pérez-Enciso, M. Assessment of genetic variation explained by markers for wool traits in sheep via a segment mapping approach. Mamm. Genome 2001, 12, 569–572. [Google Scholar] [CrossRef]
- Phua, S.H.; Dodds, K.G.; Morris, C.A.; Henry, H.M.; Beattie, A.E.; Garmonsway, H.G.; Towers, N.R.; Crawford, A.M. A genome-screen experiment to detect quantitative trait loci affecting resistance to facial eczema disease in sheep. Anim. Genet. 2008, 40, 73–79. [Google Scholar] [CrossRef]
- Atlija, M.; Arranz, J.J.; Martinez-Valladares, M.; Gutiérrez-Gil, B. Detection and replication of QTL underlying resistance to gastrointestinal nematodes in adult sheep using the ovine 50K SNP array. Genet. Sel. Evol. 2016, 48, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Alqudah, A.M.; Sallam, A.; Stephen Baenziger, P.; Börner, A. GWAS: Fast-Forwarding Gene Identification in Temperate Cereals: Barley as a Case Study—A review. J. Adv. Res. 2019, 22, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Platt, A.; Vilhjálmsson, B.J.; Nordborg, M. Conditions under which genome-wide association studies will be positively misleading. Genetics 2010, 186, 1045–1052. [Google Scholar] [CrossRef] [Green Version]
- Lôbo, A.M.B.O.; Lôbo, R.N.B.; Paiva, S.R.; De Oliveira, S.M.P.; Facó, O. Genetic parameters for growth, reproductive and maternal traits in a multibreed meat sheep population. Genet. Mol. Biol. 2009, 32, 761–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaler, A.S.; Purcell, L.C. Estimation of a significance threshold for genome-wide association studies. BMC Genom. 2019, 20, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Georgiopoulos, G.; Evangelou, E. Power considerations for λ inflation factor in meta-analyses of genome-wide association studies. Genet. Res. 2016, 98, 1–14. [Google Scholar] [CrossRef] [Green Version]
- England, B.G.; Webb, R.; Dahmer, M.K. Follicular Steroidogenesis and Gonadotropin Binding to Ovine Follicles during the Estrous Cycle. Endocrinology 1981, 109, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Murphy, B.D.; de Alba, M.J.; Manns, J.G. Endocrinology of the postpartum period in the pelibuey ewe. J. Anim. Sci. 1987, 64, 1717–1724. [Google Scholar] [CrossRef]
- Haldar, A.; Pal, P.; Datta, M.; Paul, R.; Pal, S.K.; Majumdar, D. Prolificacy and Its Relationship with Age, Body Weight, Parity, Previous Litter Size and Body Linear Type Traits in Meat-type Goats. Asian-Australas. J. Anim. Sci. 2014, 27, 628–634. [Google Scholar] [CrossRef]
- Abecia, J.A.; Palacios, C. Ewes giving birth to female lambs produce more milk than ewes giving birth to male lambs. Ital. J. Anim. Sci. 2018, 17, 736–739. [Google Scholar] [CrossRef]
- Hinch, G.N. The Sucking Behaviour of Triplet, Twin and Single Lambs at Pasture. Appl. Anim. Behav. Sci. 1989, 22, 39–48. [Google Scholar] [CrossRef]
- Constantinou, A. Genetic and Environmental Relationships of Body Weight, Milk Yield and Litter Size in Damascus Goats. Small Rumin. Res. 1989, 2, 163–174. [Google Scholar] [CrossRef]
- Martinez-royo, A.; Alabart, L.; Sarto, P.; Serrano, M.; Lahoz, B.; Folch, J.; Hugo, J. Theriogenology Genome-wide association studies for reproductive seasonality traits in Rasa Aragonesa sheep breed. Theriogenology 2017, 99, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, I.; Kawamura, K. Systems Biology in Reproductive Medicine Regulation of follicle growth through hormonal factors and mechanical cues mediated by Hippo signaling pathway. Syst. Biol. Reprod. Med. 2018, 64, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.; Li, X.; Zheng, T.; Hu, C.; Pan, Z.; Huang, J.; Li, J.; Li, W.; Zheng, Y. The Hippo Signaling Pathway Regulates Ovarian Function via the Proliferation of Ovarian Germline Stem Cells. Cell. Physiol. Biochem. 2017, 41, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Plewes, M.R.; Hou, X.; Zhang, P.; Liang, A.; Hua, G.; Wood, J.R.; Cupp, A.S.; Lv, X.; Wang, C.; Davis, J.S. Yes-associated protein 1 is required for proliferation and function of bovine granulosa cells in vitro. Biol. Reprod. 2019, 101, 1001–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harvey, K.F.; Hariharan, I.K. The Hippo Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Krapivinsky, G.; Krapivinsky, L.; Renthal, N.E.; Santa-cruz, A.; Manasian, Y.; Clapham, D.E. Histone phosphorylation by TRPM6’s cleaved kinase attenuates adjacent arginine methylation to regulate gene expression. Proc. Natl. Acad. Sci. USA 2017, 114, E7092–E7100. [Google Scholar] [CrossRef] [Green Version]
- Komiya, Y.; Runnels, L.W. TRPM Channels and Magnesium in Early Embryonic Development. Int. J. Dev. Biol. 2015, 59, 281–288. [Google Scholar] [CrossRef]
- Grummer, R.R.; Carroll, D.J. A review of lipoprotein cholesterol metabolism: Importance to ovarian function. J. Anita. Sci. 2018, 66, 3160–3173. [Google Scholar] [CrossRef]
- Nelson, S.B.; Eraly, S.A.; Mellon, P.L. The GnRH promoter: Target of transcription factors, hormones, and signaling pathways. Mol. Cell. Endocrinol. 1998, 140, 151–155. [Google Scholar] [CrossRef]
- Singh, P.; Krishna, A. Effects of GnRH agonist treatment on steroidogenesis and folliculogenesis in the ovary of cyclic mice. J. Ovarian Res. 2010, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radhakrishnan, A.; Raju, R.; Tuladhar, N.; Subbannayya, T.; Thomas, J.K.; Goel, R.; Telikicherla, D.; Palapetta, S.M.; Rahiman, B.A.; Venkatesh, D.D.; et al. A pathway map of prolactin signaling. J. Cell Commun. Signal. 2012, 6, 169–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyga, S.; Volpe, S.; Werthmann, R.C.; Götz, K.; Sungkaworn, T.; Lohse, M.J.; Calebiro, D. Persistent cAMP Signaling by Internalized LH Receptors in Ovarian Follicles. Endocrinology 2016, 157, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Lipina, T.V.; Prasad, T.; Yokomaku, D.; Luo, L.; Connor, S.A.; Kawabe, H.; Wang, Y.T.; Brose, N.; Roder, J.C.; Craig, A.M. Cognitive Deficits in Calsyntenin-2-deficient Mice Associated with Reduced GABAergic Transmission. Neuropsychopharmacology 2016, 41, 802–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, E.J.; Park, S.H.; Choi, K.C.; Leung, P.C.K.; Jeung, E.B. Identification of estrogen-regulated genes by microarray analysis of the uterus of immature rats exposed to endocrine disrupting chemicals. Reprod. Biol. Endocrinol. 2006, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Santana, M.H.A.; Ventura, R.V.; Utsunomiya, Y.T.; Neves, H.H.R.; Alexandre, P.A.; Oliveira Junior, G.A.; Gomes, R.C.; Bonin, M.N.; Coutinho, L.L.; Garcia, J.F.; et al. A genomewide association mapping study using ultrasound-scanned information identifies potential genomic regions and candidate genes affecting carcass traits in Nellore cattle. J. Anim. Breed. Genet. 2015, 132, 420–427. [Google Scholar] [CrossRef]
- Pesántez-Pacheco, J.L.; Heras-Molina, A.; Torres-Rovira, L.; Sanz-Fernández, M.V.; García-Contreras, C.; Vázquez-Gómez, M.; Feyjoo, P.; Cáceres, E.; Frías-Mateo, M.; Hernández, F.; et al. Influence of maternal factors (Weight, body condition, parity, and pregnancy rank) on plasma metabolites of dairy ewes and their lambs. Animals 2019, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Lehmana, M.N.; Ladhaa, Z.; Coolena, L.M.; Hilemanb, S.M.; Connorsb, J.M.; Goodmanb, R.L. Neuronal plasticity and seasonal reproduction in sheep. Eur. J. Neurosci. 2010, 23, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Hnia, K.; Vaccari, I.; Bolino, A.; Laporte, J. Myotubularin phosphoinositide phosphatases: Cellular functions and disease pathophysiology. Trends Mol. Med. 2012, 18, 317–327. [Google Scholar] [CrossRef]
- Chojnowski, A.; Ravisé, N.; Bachelin, C.; Depienne, C.; Ruberg, M.; Brugg, B.; Laporte, J.; Evercooren, A.B.; Leguern, E. Silencing of the Charcot–Marie–Tooth associated MTMR2 gene decreases proliferation and enhances cell death in primary cultures of Schwann cells. Neurobiol. Dis. 2007, 26, 323–331. [Google Scholar] [CrossRef]
- Sosa-Madrid, B.S.; Hernández, P.; Blasco, A.; Haley, C.S.; Fontanesi, L.; Santacreu, M.A.; Pena, R.N.; Navarro, P.; Ibáñez-Escriche, N. Genomic regions influencing intramuscular fat in divergently selected rabbit lines. Anim. Genet. 2020, 51, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mruk, D.D.; Cheng, C.Y. The myotubularin family of lipid phosphatases in disease and in spermatogenesis. Biochem. J. 2011, 176, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Wu, Y.; Su, W.; Xing, L.Y.; Shen, Y.; He, X.; Li, L.; Yuan, Y.; Tang, X.; Chen, G. 17β-estradiol enhances schwann cell differentiation via the ERβ-ERK1/2 signaling pathway and promotes remyelination in injured sciatic nerves. Front. Pharmacol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volodarsky, M.; Lichtig, H.; Leibson, T.; Sadaka, Y.; Kadir, R.; Perez, Y.; Liani-leibson, K.; Gradstein, L.; Shaco-levy, R.; Shorer, Z.; et al. CDC174, a novel component of the exon junction complex whose mutation underlies a syndrome of hypotonia and psychomotor developmental delay. Hum. Mol. Genet. 2015, 24, 6485–6491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, S.; Bick, J.; Ulbrich, S.E.; Bauersachs, S. Cell type-specific analysis of transcriptome changes in the porcine endometrium on Day 12 of pregnancy. BMC Genom. 2018, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Scholz-Romero, K.; Perez, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E.; Salomon, C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J. Transl. Med. 2014, 12, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Simmons, H.M.; Ruis, B.L.; Kapoor, M.; Hudacek, A.W.; Conklin, K.F. Identification of NOM1, a nucleolar, eIF4A binding protein encoded within the chromosome 7q36 breakpoint region targeted in cases of pediatric acute myeloid leukemia. Gene 2005, 347, 137–145. [Google Scholar] [CrossRef]
- Solomon-zemler, R.; Pozniak, Y.; Geiger, T.; Werner, H. Identification of nucleolar protein NOM1 as a novel nuclear IGF1R-interacting protein. Mol. Genet. Metab. 2019, 126, 259–265. [Google Scholar] [CrossRef]
- Gunawardena, S.R.; Ruis, B.L.; Meyer, J.A.; Kapoor, M.; Conklin, K.F. NOM1 Targets Protein Phosphatase I to the Nucleolus. J. Biol. Chem. 2008, 283, 398–404. [Google Scholar] [CrossRef] [Green Version]
- Peñagaricano, F.; Wang, X.; Rosa, G.J.M.; Radunz, A.E.; Khatib, H. Maternal nutrition induces gene expression changes in fetal muscle and adipose tissues in sheep. BMC Genet. 2014, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Marziali, F.; Dizanzo, M.P.; Cavatorta, A.L.; Gardiol, D. Differential expression of DLG1 as a common trait in different human diseases: An encouraging issue in molecular pathology. Biol. Chem. 2018, 400, 699–710. [Google Scholar] [CrossRef] [Green Version]
- Cavatorta, A.L.; Di Gregorio, A.; Valdano, M.B.; Marziali, F.; Cabral, M.; Bottai, H.; Cittadini, J.; Nocito, A.L.; Gardiol, D. DLG1 polarity protein expression associates with the disease progress of low-grade cervical intraepithelial lesions. Exp. Mol. Pathol. 2017, 102, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.H.Y.; Rajkovic, A.; Szafranski, P.; Ochsner, S.; Richards, J.; Goode, S. Expression of Drosophila neoplastic tumor suppressor genes discslarge, scribble, and lethal giant larvae in the mammalian ovary. Gene Expr. Patterns 2003, 3, 3–11. [Google Scholar] [CrossRef]
- Middelbeek, J.; Clark, K.; Leeuwen, F.N.V. The alpha-kinase family: An exceptional branch on the protein kinase tree. Cell. Mol. Life Sci. 2010, 67, 875–890. [Google Scholar] [CrossRef] [Green Version]
- Jaouadi, H.; Kraoua, L.; Chaker, L.; Atkinson, A.; Benkhalifa, R.; Mrad, R.; Abdelhak, S.; Zaffran, S. Novel ALPK3 mutation in a Tunisian patient with pediatric cardiomyopathy and facio-thoraco-skeletal features. J. Hum. Genet. 2018, 63, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, V.; Mambetisaeva, E.; William, A.; Annan, A.; Knöll, B.; Guy, T.; Lawrence, B. Dynamic Expression Patterns of Robo (Robo1 and Robo2) in the Developing Murine Central Nervous System. J. Comp. Neurol. 2004, 468, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, R.E.; Hryhorskyj, L.; Tremewan, H.; Hogg, K.; Thomson, A.A.; McNeilly, A.S.; Duncan, W.C. Europe PMC Funders Group Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction 2010, 139, 395–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, R.E.; Duncan, W.C.; Europe PMC Funders Group. The SLIT/ROBO pathway: A regulator of cell function with implications for the reproductive system. Reproduction 2010, 139, 697–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamalvandi, M.; Motovali-bashi, M.; Amirmahani, F.; Parisa, D.; Goharrizi, J.K. Association of T/A polymorphism in miR-1302 binding site in CGA gene with male infertility in Isfahan population. Mol. Biol. Rep. 2018, 45, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Quan, Y.; Zhang, M.; Wang, K.; Zhu, M.; Chen, Y.; Li, Q. Effects of pituitary-specific overexpression of FSH α/β on reproductive traits in transgenic boars. J. Anim. Sci. Biotechnol. 2017, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcdonald, R.; Sadler, C.; Kumar, T.R. Gain–of–Function Genetic Models to Study FSH Action. Front. Endocrinol. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ulloa-aguirre, A.; Dias, J.A.; Bousfield, G.R. Gonadotropins. In Endocrinology of the Testis and Male Reproduction; Simoni, M., Huhtaniem, I.T., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–52. [Google Scholar]
- Sankar, A.; Kooistra, S.M.; Gonzalez, J.M.; Ohlsson, C.; Poutanen, M.; Helin, K. Maternal expression of the histone demethylase Kdm4a is crucial for pre-implantation development. Development 2017, 144, 3264–3277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| SNP ID | Chr | Position (bp) | Gene Name | Gene Description | Traits | Signal Pathway |
|---|---|---|---|---|---|---|
| s71757.1 | 1 | 51,963,826 | ST6GALNAC3 | ST6 N-acetylgalactosaminide alpha-2 6-sialyltransferase 3 | MUSWT, LMYP, BONE_WT, BONEP, FATP [36] | Glycosphingolipid biosynthesis |
| OAR1_155672687.1 | 1 | 189,855,910 | ROBO2 | Roundabout guidance receptor 2 | MUSWT, LMYP, BONE_WT, BONEP, FATP [36] | Axon guidance |
| OAR1_204970872.1 | 1 | 189,855,910 | DLG1 | Discs large MAGUK scaffold protein 1 | ASREP [37], SAOS [43], LMYP, FATP, BONE_WT, MUSWT [36] | Hippo signaling pathway; tight junction; T-cell receptor signaling pathway |
| s09883.1 | 1 | 246,913,454 | CLSTN2 | Calsyntenin 2 | ASREP [37], TFEC_1 [44], FATP [36], FCURV [45] | |
| OAR2_65914681.1 | 2 | 61,498,071 | TRPM6 | Transient receptor potential cation channel subfamily M member 6 | HCWT [46], MF [39], MFY, PP, MY [38], BW [36] | Mineral absorption |
| OAR2_95966123.1 | 2 | 89,499,669 | COL11A1 | Collagen type XI alpha 1 chain | SCS [46], PP [38], LATRICH2 [47], HCWT [36], MF [38] | Protein digestion and absorption |
| OAR3_85112203.1 | 3 | 80,398,784 | ABCG5 | ATP-binding cassette subfamily G member 5 | INTFAT [36], MCLA [48], SL [49] | ABC transporters; fat digestion and absorption; bile secretion; cholesterol metabolism |
| OAR3_104545117_X.1 | 3 | 98,126,615 | HTRA2 | HtrA serine peptidase 2 | FECZ [50], MCLA [48], SL [49], INTFAT [36] | Apoptosis |
| s62827.1 | 8 | 49,878,423 | CGA | Glycoprotein hormones alpha polypeptide | LATRICH2 [47], INTFAT [36], FECGEN [51] | cAMP signaling pathway; GnRH signaling pathway; ovarian steroidogenesis; prolactin signaling pathway; thyroid hormone synthesis; regulation of lipolysis in adipocytes |
| OAR8_53593379.1 | 8 | 49,981,252 | HTR1E | 5-hydroxytryptamine receptor 1E | LATRICH2 [47], INTFAT [36], FECGEN [51] | cAMP signaling pathway; neuroactive ligand–receptor interaction; serotonergic synapse; taste transduction |
| OAR9_36598045.1 | 9 | 34,604,487 | ATP6V1H | ATPase H+ transporting V1 subunit H | HCWT, LMA [36] | Oxidative phosphorylation; metabolic pathways; phagosome; mTOR signaling pathway; synaptic vesicle cycle |
| OAR15_13905772.1 | 15 | 13,872,637 | MTMR2 | Myotubularin related protein 2 | Inositol phosphate metabolism; metabolic pathways; phosphatidylinositol signaling system | |
| s07255.1 | 23 | 47,438,785 | ST8SIA5 | ST8 alpha-N-acetyl-neuraminide alpha-2 8-sialyltransferase 5 | IGA [51], FATP, LMYP, HCWT, BW, FATWT [36], MFY, MY [38] | Glycosphingolipid biosynthesis; metabolic pathways |
| s08197.1 | 25 | 40,382,673 | GRID1 | Glutamate ionotropic receptor delta type subunit 1 | SL, MFDIAM, CVFD_PRI [49] | Neuroactive ligand–receptor interaction |
| Chr | SNP | Position (bp) | A1 | A2 | F_A | F_U | MAF | p-Unadjusted | Chis-q | Gene Annotated |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | s09883.1 | 246,913,454 | A | G | 0.1458 | 0.587 | 0.3617 | 8.61 × 10−6 | 19.8 | CLSTN2 |
| 15 | OAR15_13905772.1 | 13,872,637 | A | G | 0.04167 | 0.3261 | 0.1809 | 0.0003417 | 12.83 | MTMR2 |
| 19 | s15631.1 | 57,489,437 | A | C | 0.5208 | 0.1739 | 0.3511 | 0.0004272 | 12.41 | CCDC174 |
| 4 | s37914.1 | 117,719,020 | G | A | 0.6667 | 0.3043 | 0.4894 | 0.0004434 | 12.34 | NOM1 |
| 14 | s57545.1 | 13,903,063 | T | C | 0.5417 | 0.1957 | 0.3723 | 0.0005225 | 12.03 | ANKRD11 |
| 1 | OAR1_204970872.1 | 189,855,910 | T | C | 0.3542 | 0.06522 | 0.2128 | 0.0006221 | 11.71 | DLG1 |
| 18 | OAR18_22964031.1 | 22,346,018 | C | T | 0.1875 | 0.5217 | 0.3511 | 0.0006891 | 11.52 | ALPK3 |
| 1 | OAR1_155672687.1 | 144,029,243 | C | T | 0.625 | 0.2826 | 0.4574 | 0.0008655 | 11.1 | ROBO2 |
| 8 | s62827.1 | 49,878,423 | A | G | 0.08333 | 0.3696 | 0.2234 | 0.0008669 | 11.09 | CGA |
| 1 | OAR1_18691972.1 | 18,481,816 | T | C | 0.25 | 0.587 | 0.4149 | 0.0009179 | 10.99 | KDM4A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Montiel, W.; Martínez-Núñez, M.A.; Ramón-Ugalde, J.P.; Román-Ponce, S.I.; Calderón-Chagoya, R.; Zamora-Bustillos, R. Genome-Wide Association Study Reveals Candidate Genes for Litter Size Traits in Pelibuey Sheep. Animals 2020, 10, 434. https://doi.org/10.3390/ani10030434
Hernández-Montiel W, Martínez-Núñez MA, Ramón-Ugalde JP, Román-Ponce SI, Calderón-Chagoya R, Zamora-Bustillos R. Genome-Wide Association Study Reveals Candidate Genes for Litter Size Traits in Pelibuey Sheep. Animals. 2020; 10(3):434. https://doi.org/10.3390/ani10030434
Chicago/Turabian StyleHernández-Montiel, Wilber, Mario Alberto Martínez-Núñez, Julio Porfirio Ramón-Ugalde, Sergio Iván Román-Ponce, Rene Calderón-Chagoya, and Roberto Zamora-Bustillos. 2020. "Genome-Wide Association Study Reveals Candidate Genes for Litter Size Traits in Pelibuey Sheep" Animals 10, no. 3: 434. https://doi.org/10.3390/ani10030434
APA StyleHernández-Montiel, W., Martínez-Núñez, M. A., Ramón-Ugalde, J. P., Román-Ponce, S. I., Calderón-Chagoya, R., & Zamora-Bustillos, R. (2020). Genome-Wide Association Study Reveals Candidate Genes for Litter Size Traits in Pelibuey Sheep. Animals, 10(3), 434. https://doi.org/10.3390/ani10030434





