Identification and Profiling of Pituitary microRNAs of Sheep during Anestrus and Estrus Stages
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample Collection and Preparation
2.3. Total RNA Extraction and Small RNA Sequencing
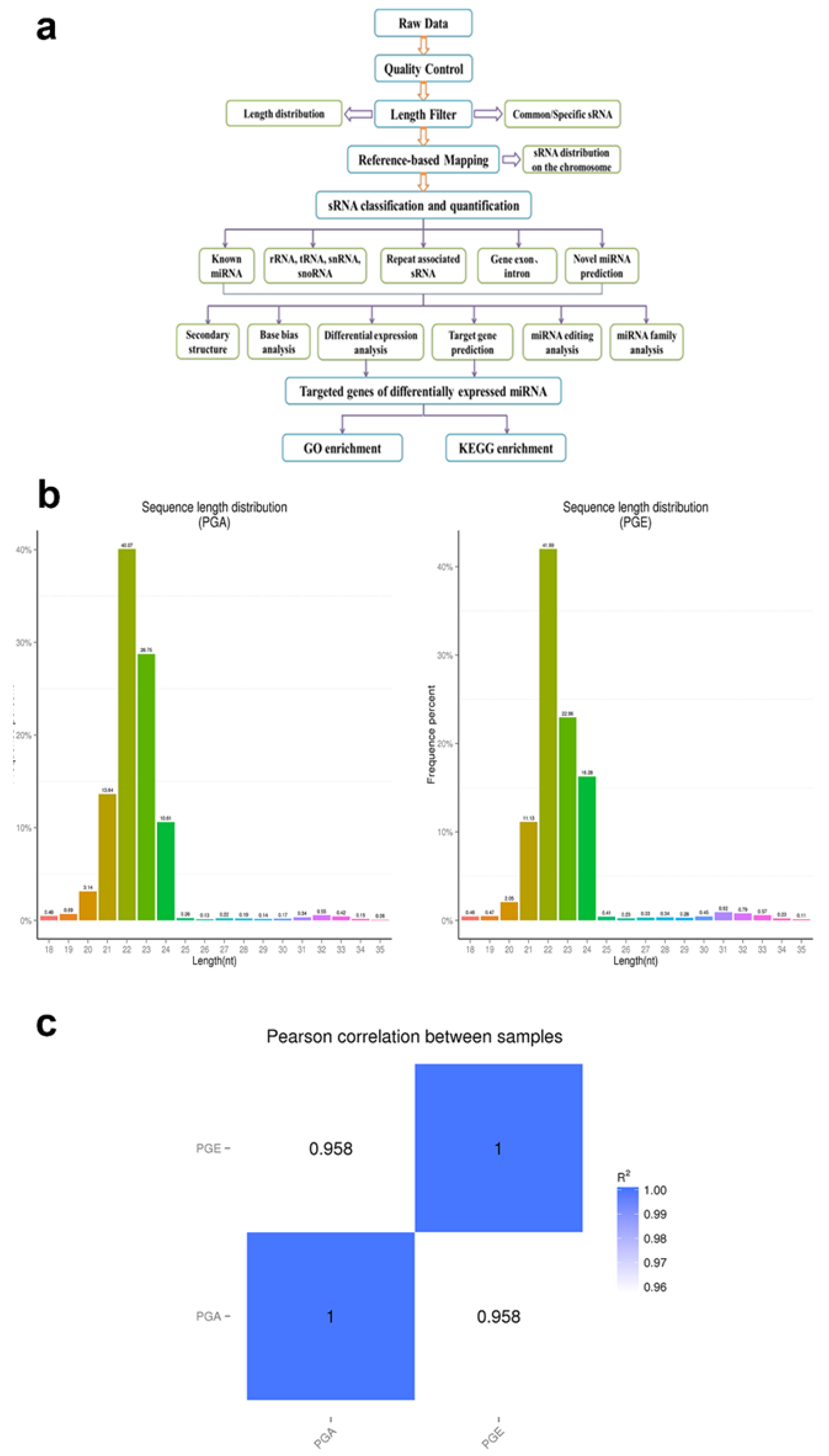
2.4. Bioinformatics Analysis of Small RNA Sequences
2.5. Real-Time Quantitative Reverse Transcription PCR Analysis
2.6. Gene Prediction and Target Enrichment Analysis
3. Results
3.1. Deep Sequencing of Sheep Pituitary Small RNA
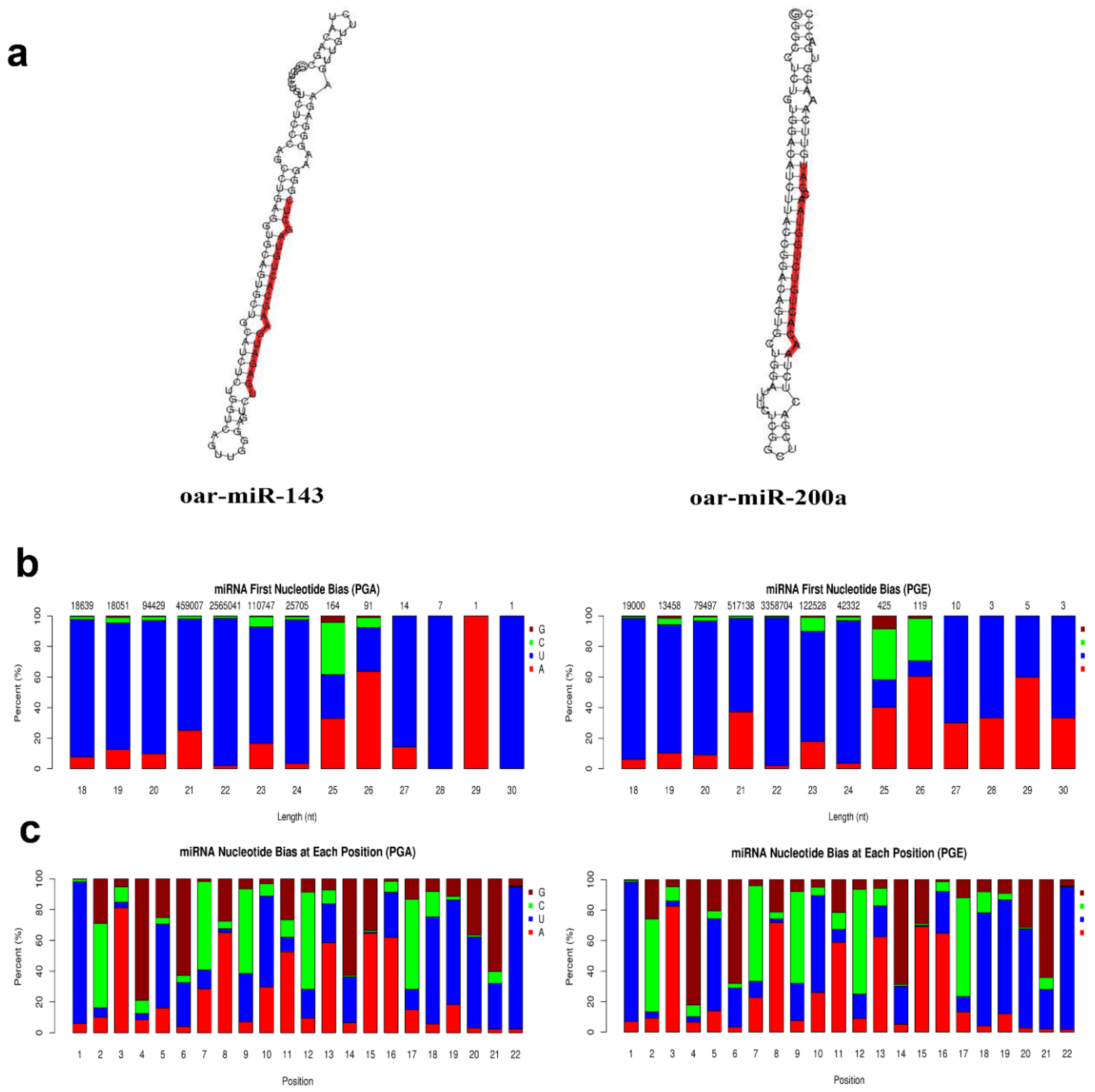
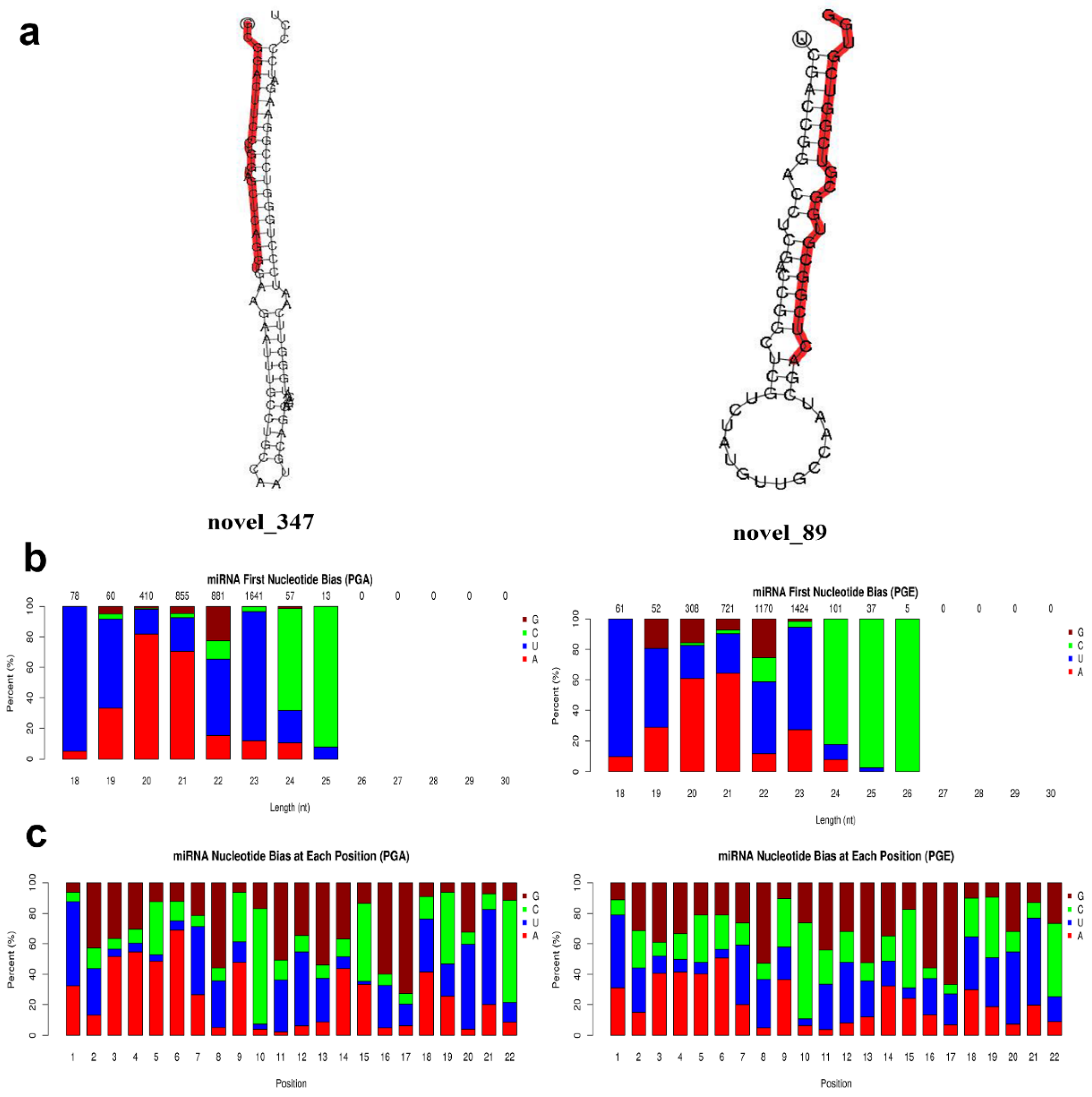
3.2. Analysis of Known and Novel miRNAs
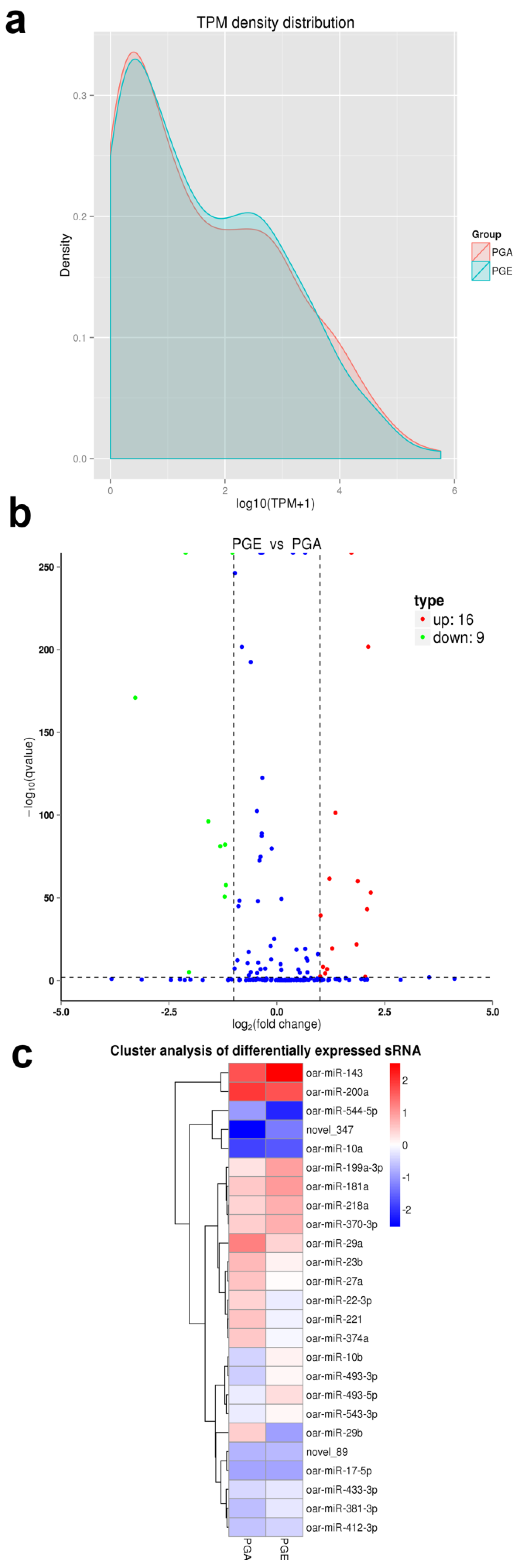
3.3. Differential Analysis of Sheep Pituitary miRNAs
3.4. Confirmation of Differentially Expressed miRNAs
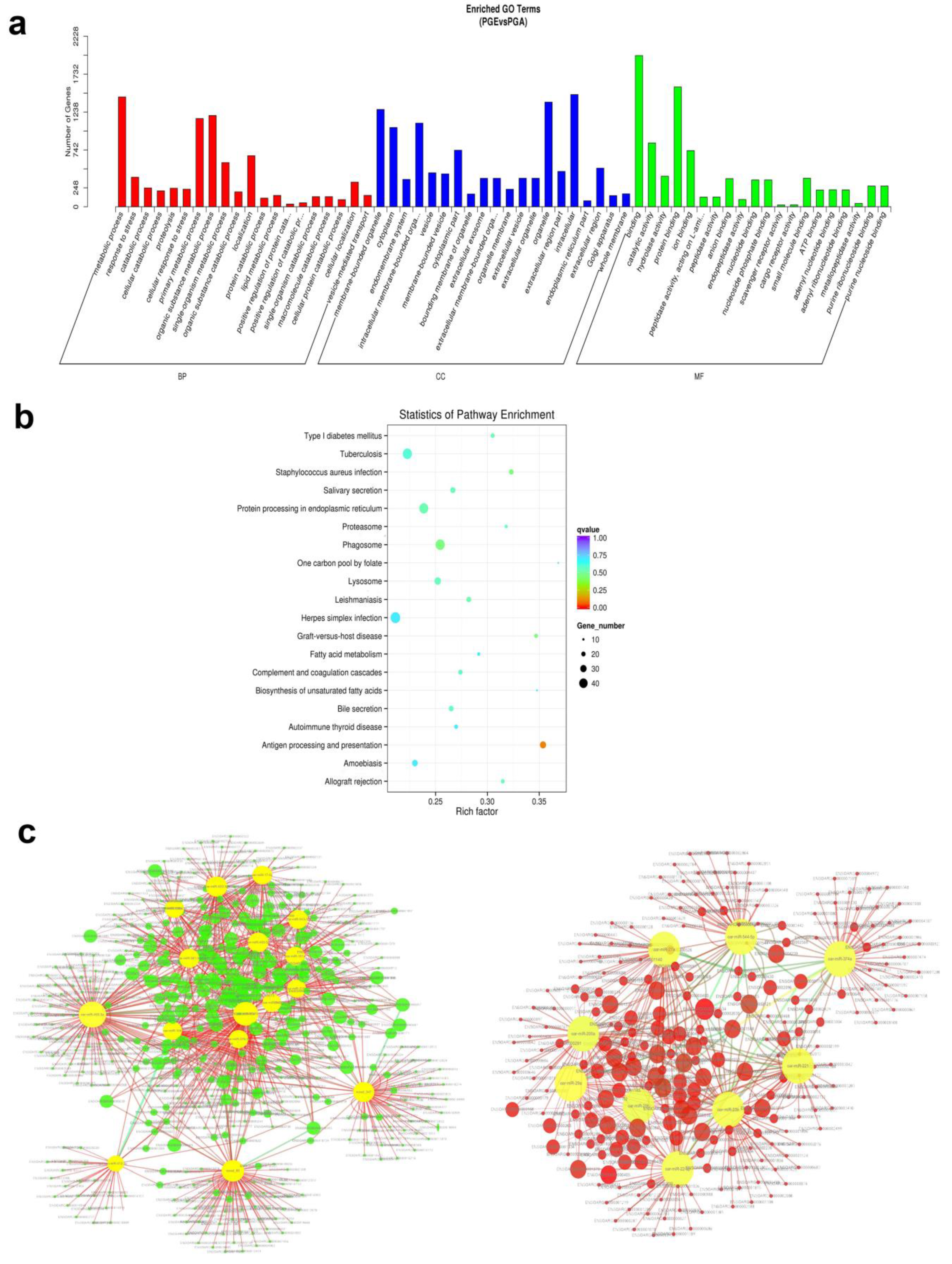
3.5. Target Prediction and Pathways Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- León, L.E.; Calligaris, S.D. Visualization and Analysis of MiRNA–Targets Interactions Networks. Methods Mol. Biol. 2017, 1509, 4939–6524. [Google Scholar]
- Sontheimer, E.J. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005, 6, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Yang, H.; Lin, S.; Lei, X.; Yuan, C.; Tian, Z.; Yu, Y.; Zhao, Z.; Chen, J. Identification and profiling of microRNAs from ovary of estrous Kazakh sheep induced by nutritional status in the anestrous season. Anim. Reprod. Sci. 2016, 175, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Ruvkun, G. Glimpses of a tiny RNA world. Science 2001, 294, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Hansen, J.H.; Hedegaard, J.; Nielsen, R.O.; Panitz, F.; Bendixen, C.; Thomsen, B. MicroRNA identity and abundance in porcine skeletal muscles determined by deep sequencing. Anim. Genet. 2010, 41, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 2003, 113, 673–676. [Google Scholar] [CrossRef]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, J.W.; Kim, S. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, R.; Xu, J.; Wu, C.I.; Lu, X. Functional conservation of both CDS- and 3′-UTR-located microRNA binding sites between species. Mol. Biol. Evol. 2015, 32, 623–628. [Google Scholar] [CrossRef]
- Zhou, H.; Rigoutsos, I. MiR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA 2014, 20, 1431–1439. [Google Scholar] [CrossRef]
- Li, H.; Xi, Q.; Xiong, Y.; Cheng, X.; Qi, Q.; Yang, L.; Shu, G.; Wang, S.; Wang, L.; Gao, P.; et al. A comprehensive expression profile of microRNAs in porcine pituitary. PLoS ONE 2011, 6, e24883. [Google Scholar] [CrossRef] [PubMed]
- Zichao, Z.; Sergio, F.; Arthur, G.H.; James, F.M.; Brad, A.A. MicroRNAs Regulate Pituitary Development, and MicroRNA 26b Specifically Targets Lymphoid Enhancer Factor 1 (Lef-1), Which Modulates Pituitary Transcription Factor 1 (Pit-1) Expression. J. Biol. Chem. 2010, 285, 34718–34728. [Google Scholar]
- De Gier, J.; Beijerink, N.J.; Kooistra, H.S.; Okkens, A.C. Physiology of the Canine Anoestrus and Methods for Manipulation of Its Length. Authors. J. Compil. 2008, 43, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Mak, T.W.; Saunders, M.E. The Immune Response Basic and Clinical Principles; The Humoral Response: B Cell Development and Activation; Elsevier Health Sciences: Amsterdam, The Netherlands, 2006; pp. 209–245. ISBN 978-0-12-088451-3. [Google Scholar]
- Nemoto, T.; Mano, A.; Shibasaki, T. Increased expression of miR-325-3p by urocortin 2 and its involvement in stress-induced suppression of LH secretion in rat pituitary. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E781–E787. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.E.; Wondisford, F.E.; Radovick, S. Role of Pit-1 in the gene expression of growth hormone, prolactin, and thyrotropin. Endocrinol. Metab. Clin. N. Am. 1996, 3, 523–540. [Google Scholar] [CrossRef]
- Zhang, N.; Lin, J.K.; Chen, J.; Liu, X.F.; Liu, J.L.; Luo, H.S.; Li, Y.Q.; Cui, S. MicroRNA 375 mediates the signaling pathway of corticotropin-releasing factor (CRF) regulating pro-opiomelanocortin (POMC) expression by targeting mitogen-activated protein kinase 8. J. Biol. Chem. 2013, 288, 10361–10373. [Google Scholar] [CrossRef]
- Hou, L.; Ji, Z.; Wang, G.; Wang, J.; Chao, T.; Wang, J. Identification and characterization of micoRNAs in the intestinal tissues of sheep. PLoS ONE 2018, 13, e0193371. [Google Scholar] [CrossRef]
- Wong, L.L.; Rademaker, M.T.; Saw, E.L.; Lew, K.S.; Ellmers, L.J.; Charles, C.J.; Richards, A.M.; Wang, P. Identification of novel microRNAs in the sheep heart and their regulation in heart failure. Sci. Rep. 2017, 7, 8250. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Ni, W.; Wang, D.; Hou, X.; Liu, Z.; Cao, Y.; Yao, Y.; Zhang, X.; Hu, S. Identification and comparison of microRNAs in pituitary gland during prenatal and post natal stages of sheep by deep sequencing. J. Genet. 2018, 97, 965–975. [Google Scholar] [CrossRef]
- Hanif, Q.; Farooq, M.; Amin, I.; Mansoor, S.; Zhang, Y.; Khan, Q.M. In silico identification of conserved miRNAs and their selective target gene prediction inn in indicine (Bos indicus) cattle. PLoS ONE 2018, 13, e0206154. [Google Scholar] [CrossRef]
- Liao, R.; Lv, Y.; Zhu, L.; Lin, Y. Altered expression of miRNAs and mRNAs reveals the potential regulatory role of miRNAs in the developmental process of early weaned goats. PLoS ONE 2019, 14, e0220907. [Google Scholar] [CrossRef] [PubMed]
- Forcada, F.; Abecia, J.A. The effect of nutrition on the seasonality of reproduction in ewes. Reprod. Nutr. Dev. 2006, 46, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Geoffrey, M.; Joshua, M.T.; Brent, D.J. Ovulatory cycle effects on tip earnings by lap dancers: Economic evidence for human estrus. Evol. Hum. Behav. 2007, 28, 375–381. [Google Scholar]
- Robert, S.Y.; Walter, R.T. Clinical Aspects of Seasonality in Mares. In Current Therapy in Large Animal Theriogenology, 2nd ed.; Gary, N., Daniel, C.S., Gillian, R., Brian, D.C., Michael, B.P., Eds.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2006; pp. 68–73. ISBN 9781437713404. [Google Scholar]
- Li, X.; Li, C.; Wei, J.; Ni, W.; Xu, Y.; Yao, R.; Zhang, M.; Li, H.; Liu, L.; Dang, H.; et al. Comprehensive Expression Profiling Analysis of Pituitary Indicates that circRNA Participates in the Regulation of Sheep Estrus. Genes 2019, 10, 90. [Google Scholar] [CrossRef]
- Zhai, M.; Xie, Y.; Liang, H.; Lei, X.; Zhao, Z. Comparative profiling of differentially expressed microRNAs in estrous ovaries of Kazakh sheep in different seasons. Gene 2018, 664, 181–191. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated profiling of microRNAs and mRNAs: Micrornas located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Carletti, M.Z.; Fiedler, S.D.; Christenson, L.K. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol. Reprod. 2010, 83, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Donadeu, F.X.; Schauer, S.N.; Sontakke, S.D. Involvement of miRNAs in ovarian follicular and luteal development. J. Endocrinol. 2012, 215, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.S.; Li, M.; Qi, Q.E.; Cheng, X.; Chen, T.; Li, C.Y.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; et al. Comparative Anterior Pituitary miRNA and mRNA Expression Profiles of Bama Mini pigs and Landrace Pigs Reveal Potential Molecular Network Involved in Animal Postnatal Growth. PLoS ONE 2015, 10, e0131987. [Google Scholar]
- Miao, X.; Luo, Q.; Qin, X.; Guo, Y. Genome wide analysis of microRNAs identifies the lipid metabolism pathway to be a defining factor in adipose tissue from different sheep. Sci. Rep. 2015, 5, 18470. [Google Scholar] [CrossRef]
- Hasuwa, H.; Ueda, J.; Ikawa, M.; Okabe, M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 2013, 341, 71–73. [Google Scholar] [CrossRef]
- Lehmann, W.; Mossmann, D.; Kleemann, J.; Mock, K.; Meisinger, C.; Brummer, T.; Herr, R.; Brabletz, S.; Stemmler, M.P.; Brabletz, T. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat. Commun. 2016, 7, 10498. [Google Scholar] [CrossRef]
- Funes, S.; Hedrick, J.A.; Vassileva, G.; Markowitz, L.; Abbondanzo, S.; Golovko, A.; Yang, S.; Monsma, F.J.; Gustafson, E.L. The KiSS-1 receptor GPR54 is essential for the development of the murine reproductive system. Biochem. Biophys. Res. Commun. 2003, 312, 1357–1363. [Google Scholar] [CrossRef]
- Muir, A.I.; Chamberlain, L.; Elshourbagy, N.A.; Michalovich, D.; Moore, D.J.; Calamari, A.; Szekeres, P.G.; Sarau, H.M.; Chambers, J.K.; Murdock, P.; et al. A novel human G protein-coupled receptor, activated by the peptide KiSS-1. J. Biol. Chem. 2001, 276, 28969–28975. [Google Scholar] [CrossRef]
- Navarro, V.M.; Castellano, J.M.; Fernández-Fernández, R.; Barreiro, M.L.; Roa, J.; Sanchez-Criado, J.E.; Aguilar, E.; Dieguez, C.; Pinilla, L.; Tena-Sempere, M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology 2004, 145, 4565–4574. [Google Scholar] [CrossRef]
- Arzt, E. gp130 cytokine signaling in the pituitary gland: A paradigm for cytokine–neuro-endocrine pathways. J. Clin. Investig. 2001, 108, 1729–1733. [Google Scholar] [CrossRef] [PubMed]
- Haston, S.; Pozzi, S.; Carreno, G.; Manshaei, S.; Panousopoulos, L.; Gonzalez-Meljem, J.M.; Apps, J.R.; Virasami, A.; Thavaraj, S.; Gutteridge, A.; et al. MAPK pathway control of stem cell proliferation and differentiation in the embryonic pituitary provides insights into the pathogenesis of papillary craniopharyngioma. Development 2017, 144, 2141–2152. [Google Scholar] [CrossRef] [PubMed]


| Gene_ID | PGE | PGA | log2.Fold_change. | q-Value | Corrected p-Value | UP/DOWN Regulate |
|---|---|---|---|---|---|---|
| oar-miR-143 | 32670.824 | 9873.594248 | 1.7264 | 0 | 0 | UP |
| oar-miR-200a | 8144.9197 | 16627.14523 | −1.0296 | 0 | 0 | DOWN |
| oar-miR-29a | 923.39904 | 3991.937194 | −2.1121 | 0 | 0 | DOWN |
| oar-miR-199a-3p | 2190.8655 | 504.9304155 | 2.1173 | 1.02 × 10−203 | 1.69× 10−202 | UP |
| oar-miR-29b | 85.951935 | 836.0509809 | −3.282 | 9.42 × 10−173 | 1.21× 10−171 | DOWN |
| oar-miR-181a | 2319.1442 | 904.6077302 | 1.3582 | 4.40 × 10−103 | 4.58× 10−102 | UP |
| oar-miR-221 | 327.96766 | 986.8189359 | −1.5892 | 5.91 × 10−98 | 5.79× 10−97 | DOWN |
| oar-miR-23b | 520.64541 | 1197.040669 | −1.2011 | 8.90 × 10−84 | 7.41× 10−83 | DOWN |
| oar-miR-27a | 426.90327 | 1058.504832 | −1.31 | 7.82 × 10−83 | 6.20× 10−82 | DOWN |
| oar-miR-218a | 1721.6354 | 738.194044 | 1.2217 | 4.35 × 10−63 | 2.89× 10−62 | UP |
| oar-miR-493-5p | 780.5786 | 212.7819441 | 1.8752 | 1.55 × 10−61 | 9.90× 10−61 | UP |
| oar-miR-374a | 379.12334 | 858.2394724 | −1.1787 | 3.64 × 10−59 | 2.24× 10−58 | DOWN |
| oar-miR-10b | 549.72884 | 121.4677676 | 2.1781 | 1.17 × 10−54 | 6.98× 10−54 | UP |
| oar-miR-22-3p | 314.72431 | 728.5221375 | −1.2109 | 3.23 × 10−52 | 1.85× 10−51 | DOWN |
| oar-miR-493-3p | 470.78809 | 110.3735218 | 2.0927 | 1.80 × 10−44 | 8.81× 10−44 | UP |
| oar-miR-370-3p | 1608.4178 | 797.0788868 | 1.0128 | 1.16 × 10−40 | 5.52× 10−40 | UP |
| oar-miR-381-3p | 287.19891 | 79.65099512 | 1.8503 | 3.02 × 10−23 | 1.36× 10−22 | UP |
| oar-miR-543-3p | 487.66687 | 200.549827 | 1.2819 | 8.80 × 10−21 | 3.76× 10−20 | UP |
| novel_347 | 42.846131 | 0 | 6.2348 | 4.85× 10−11 | 1.58× 10−10 | UP |
| oar-miR-433-3p | 287.45859 | 136.8290309 | 1.071 | 2.14× 10−9 | 6.84× 10−9 | UP |
| oar-miR-412-3p | 201.24698 | 89.89183734 | 1.1627 | 4.97× 10−8 | 1.50× 10−7 | UP |
| oar-miR-544-5p | 8.8288997 | 36.12741564 | −2.0328 | 3.47× 10−6 | 9.64× 10−6 | DOWN |
| novel_89 | 130.61578 | 60.02271418 | 1.1217 | 2.29× 10−5 | 5.96× 10−5 | UP |
| oar-miR-17-5p | 92.443773 | 46.08379003 | 1.0043 | 0.001524 | 0.0038437 | UP |
| oar-miR-10a | 22.331923 | 5.404888954 | 2.0468 | 0.0027333 | 0.0067909 | UP |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, Y.; Li, C.; Li, X.; Ni, W.; Yao, R.; Xu, Y.; Quan, R.; Li, H.; Zhang, M.; Liu, L.; et al. Identification and Profiling of Pituitary microRNAs of Sheep during Anestrus and Estrus Stages. Animals 2020, 10, 402. https://doi.org/10.3390/ani10030402
Ullah Y, Li C, Li X, Ni W, Yao R, Xu Y, Quan R, Li H, Zhang M, Liu L, et al. Identification and Profiling of Pituitary microRNAs of Sheep during Anestrus and Estrus Stages. Animals. 2020; 10(3):402. https://doi.org/10.3390/ani10030402
Chicago/Turabian StyleUllah, Yaseen, Cunyuan Li, Xiaoyue Li, Wei Ni, Rui Yao, Yueren Xu, Renzhe Quan, Huixiang Li, Mengdan Zhang, Li Liu, and et al. 2020. "Identification and Profiling of Pituitary microRNAs of Sheep during Anestrus and Estrus Stages" Animals 10, no. 3: 402. https://doi.org/10.3390/ani10030402
APA StyleUllah, Y., Li, C., Li, X., Ni, W., Yao, R., Xu, Y., Quan, R., Li, H., Zhang, M., Liu, L., Hu, R., Guo, T., Li, Y., Wang, X., & Hu, S. (2020). Identification and Profiling of Pituitary microRNAs of Sheep during Anestrus and Estrus Stages. Animals, 10(3), 402. https://doi.org/10.3390/ani10030402





