Comparison of Two DNA Extraction Methods and Two PCRs for Detection of Echinococcus multilocularis in the Stool Samples of Naturally Infected Red Foxes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. Polymerase Chain Reactions (PCRs)
2.3. Statistical Analysis
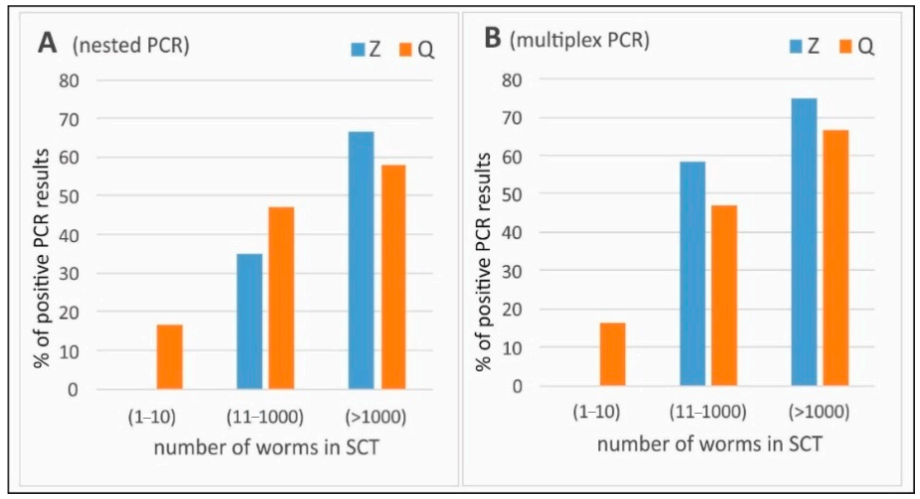
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nakao, M.; Xiao, N.; Okamoto, M.; Yanagida, T.; Sako, Y.; Ito, A. Geographic pattern of genetic variation in the fox tapeworm Echinococcus multilocularis. Parasitol. Int. 2009, 58, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, A.; Siles-Lucas, M.; Karamon, J.; Possenti, A.; Conraths, F.J.; Romig, T.; Wysocki, P.; Mannocci, A.; Mipatrini, D.; La Torre, G.; et al. The geographical distribution and prevalence of Echinococcus multilocularis in animals in the European Union and adjacent countries: A systematic review and meta-analysis. Parasites Vectors 2016, 9, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Kapel, C.M.O.; Torgerson, P.R.; Thompson, R.C.A.; Deplazes, P. Reproductive potential of Echinococcus multilocularis in experimentally infected foxes, dogs, raccoon dogs and cats. Int. J. Parasitol. 2006, 36, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Romig, T. Echinococcus multilocularis in Europe—state of the art. Vet. Res. Commun. 2009, 33, S31–S34. [Google Scholar] [CrossRef] [PubMed]
- Hegglin, D.; Deplazes, P. Control of Echinococcus multilocularis: Strategies, feasibility and cost-benefit analyses. Int. J. Parasitol. 2013, 43, 327–337. [Google Scholar] [CrossRef]
- Deplazes, P.; Alther, P.; Tanner, I.; Thompson, R.C.A.; Eckert, J. Echinococcus multilocularis coproantigen detection by enzyme-linked immunosorbent assay in fox, dog, and cat populations. J. Parasitol. 1999, 85, 115–121. [Google Scholar] [CrossRef]
- Dinkel, A.; Von Nickisch-Rosenegk, M.; Bilger, B.; Merli, M.; Lucius, R.; Romig, T. Detection of Echinococcus multilocularis in the definitive host: Coprodiagnosis by PCR as an alternative to necropsy. J. Clin. Microbiol. 1998, 36, 1871–1876. [Google Scholar] [CrossRef]
- Trachsel, D.; Deplazes, P.; Mathis, A. Identification of taeniid eggs in the faeces from carnivores based on multiplex PCR using targets in mitochondrial DNA. Parasitology 2007, 134, 911–920. [Google Scholar] [CrossRef]
- Isaksson, M.; Hagstrom, A.; Armua-Fernandez, M.T.; Wahlstrom, H.; Agren, E.O.; Miller, A.; Holmberg, A.; Lukacs, M.; Casulli, A.; Deplazes, P.; et al. A semi-automated magnetic capture probe based DNA extraction and real-time PCR method applied in the Swedish surveillance of Echinococcus multilocularis in red fox (Vulpes vulpes) faecal samples. Parasites Vectors 2014, 7, 583. [Google Scholar] [CrossRef]
- Maas, M.; Van Roon, A.; Dam-Deisz, C.; Opsteegh, M.; Massolo, A.; Deksne, G.; Teunis, P.; Van der Giessen, J. Evaluation by latent class analysis of a magnetic capture based DNA extraction followed by real-time qPCR as a new diagnostic method for detection of Echinococcus multilocularis in definitive hosts. Vet. Parasitol. 2016, 230, 20–24. [Google Scholar] [CrossRef]
- Dinkel, A.; Kern, S.; Brinker, A.; Oehme, R.; Vaniscotte, A.; Giraudoux, P.; Mackenstedt, U.; Romig, T. A real-time multiplex-nested PCR system for coprological diagnosis of Echinococcus multilocularis and host species. Parasitol. Res. 2011, 109, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Knapp, J.; Umhang, G.; Poulle, M.-L.; Millon, L. Development of a Real-Time PCR for a Sensitive One-Step Coprodiagnosis Allowing both the Identification of Carnivore Feces and the Detection of Toxocara spp. and Echinococcus multilocularis. Appl. Environ. Microbiol. 2016, 82, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Hofer, S.; Gloor, S.; Muller, U.; Mathis, A.; Hegglin, D.; Deplazes, P. High prevalence of Echinococcus multilocularis in urban red foxes (Vulpes vulpes) and voles (Arvicola terrestris) in the city of Zurich, Switzerland. Parasitology 2000, 120, 135–142. [Google Scholar] [CrossRef] [PubMed]
- OIE. Echinococcosis/hydatidosis (infection with Echinococcus granulosus and E. multilocularis). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 6th ed.; Office International des Epizooties: Paris, France, 2008; Volume 1, pp. 1–15. [Google Scholar]
- Karamon, J.; Sroka, J.; Cencek, T. The first detection of Echinococcus multilocularis in slaughtered pigs in Poland. Vet. Parasitol. 2012, 185, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Liccioli, S.; Massolo, A. Egg intensity and freeze-thawing of fecal samples affect sensitivity of Echinococcus multilocularis detection by PCR. Parasitol Res 2014, 113, 3867–3873. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, P.; Schares, G.; Press, S.; Frohlich, A.; Basso, W.; Herzig, M.; Conraths, F.J. Comparison of different commercial DNA extraction kits and PCR protocols for the detection of Echinococcus multilocularis eggs in faecal samples from foxes. Vet. Parasitol. 2017, 237, 83–93. [Google Scholar] [CrossRef]
- Karamon, J.; Sroka, J.; Cencek, T.; Kochanowski, M.; Dąbrowska, J. Efficacy of intestinal scraping technique in the detection of Echinococcus multilocularis—Estimation of the limit of the detection and comparison with sedimentation and counting technique. Bull. Vet. Inst. Pulawy 2012, 56, 535–538. [Google Scholar] [CrossRef][Green Version]
- EFSA. The EU summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Ourbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar]
- Irie, T.; Ito, T.; Kouguchi, H.; Yamano, K.; Uraguchi, K.; Yagi, K.; Nonaka, N. Diagnosis of canine Echinococcus multilocularis infections by copro-DNA tests: Comparison of DNA extraction techniques and evaluation of diagnostic deworming. Parasitol. Res. 2017, 116, 2139–2144. [Google Scholar] [CrossRef]
- Al-Sabi, M.N.S.; Kapel, C.M.O.; Deplazes, P.; Mathis, A. Comparative copro-diagnosis of Echinococcus multilocularis in experimentally infected foxes. Parasitol. Res. 2007, 101, 731–736. [Google Scholar] [CrossRef]
- Mathis, A.; Deplazes, P.; Eckert, J. An improved test system for PCR-based specific detection of Echinococcus multilocularis eggs. J. Helminthol. 1996, 70, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Karamon, J. Detection of Echinococcus multilocularis in faeces by nested PCR with the use of diluted DNA samples. Pol. J. Vet. Sci. 2014, 17, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Oines, O.; Isaksson, M.; Hagstrom, A.; Tavornpanich, S.; Davidson, R.K. Laboratory assessment of sensitive molecular tools for detection of low levels of Echinococcus multilocularis-eggs in fox (Vulpes vulpes) faeces. Parasites Vectors 2014, 7, 246. [Google Scholar] [CrossRef] [PubMed]
| Method of Isolation | Percentage of Positive Results in Echinococcus multilocularis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nested PCR | Multiplex PCR | |||||||||
| Total | 1/1 | 1/10 | Only 1/1 | Only 1/10 | Total | 1/1 | 1/10 | Only 1/1 | Only 1/10 | |
| Z | 40.0 | 40.0 | 34.3 | 5.7 | - | 54.3 | 51.4 | 42.9 | 11.4 | 2.9 |
| Q | 45.7 | 45.7 | 25.7 | 20.0 | - | 48.6 | 45.7 | 28.6 | 22.9 | 2.9 |
| Method of Isolation | Percentage of Positive Results in Echinococcus multilocularis Internal Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nested PCR | Multiplex PCR | |||||||||
| Total | 1/1 | 1/10 | Only 1/1 | Only 1/10 | Total | 1/1 | 1/10 | Only 1/1 | Only 1/10 | |
| Z | 100.0 | 97.9 a | 100.0 | 0.0 | 2.1 | 100.0 | 100.0 | 100.0 | 0.0 | 2.1 |
| Q | 97.9 | 83.3 b | 97.9 | 0.0 | 14.6 | 100.0 | 100.0 | 97.9 | 0.0 | 0.0 |
| Method of Isolation | Percentage of Positive Results for Taenia spp. | ||||
|---|---|---|---|---|---|
| Multiplex PCR | |||||
| Total | 1/1 | 1/10 | only 1/1 | only 1/10 | |
| Z | 87.5 | 87.5 | 77.1 | 10.4 | - |
| Q | 91.7 | 89.6 | 64.6 | 27.1 | 2.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skrzypek, K.; Karamon, J.; Samorek-Pieróg, M.; Dąbrowska, J.; Kochanowski, M.; Sroka, J.; Bilska-Zając, E.; Cencek, T. Comparison of Two DNA Extraction Methods and Two PCRs for Detection of Echinococcus multilocularis in the Stool Samples of Naturally Infected Red Foxes. Animals 2020, 10, 2381. https://doi.org/10.3390/ani10122381
Skrzypek K, Karamon J, Samorek-Pieróg M, Dąbrowska J, Kochanowski M, Sroka J, Bilska-Zając E, Cencek T. Comparison of Two DNA Extraction Methods and Two PCRs for Detection of Echinococcus multilocularis in the Stool Samples of Naturally Infected Red Foxes. Animals. 2020; 10(12):2381. https://doi.org/10.3390/ani10122381
Chicago/Turabian StyleSkrzypek, Katarzyna, Jacek Karamon, Małgorzata Samorek-Pieróg, Joanna Dąbrowska, Maciej Kochanowski, Jacek Sroka, Ewa Bilska-Zając, and Tomasz Cencek. 2020. "Comparison of Two DNA Extraction Methods and Two PCRs for Detection of Echinococcus multilocularis in the Stool Samples of Naturally Infected Red Foxes" Animals 10, no. 12: 2381. https://doi.org/10.3390/ani10122381
APA StyleSkrzypek, K., Karamon, J., Samorek-Pieróg, M., Dąbrowska, J., Kochanowski, M., Sroka, J., Bilska-Zając, E., & Cencek, T. (2020). Comparison of Two DNA Extraction Methods and Two PCRs for Detection of Echinococcus multilocularis in the Stool Samples of Naturally Infected Red Foxes. Animals, 10(12), 2381. https://doi.org/10.3390/ani10122381






