Exposure to Zoonotic West Nile Virus in Long-Tailed Macaques and Bats in Peninsular Malaysia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Study Design
2.3. Serological Analysis
2.4. Reverse Transcriptase—Polymerase Chain Reaction (RT-PCR) Assay
2.5. DNA Sequencing and Bioinformatic Analysis
3. Results
3.1. WNV Antibodies in Macaques
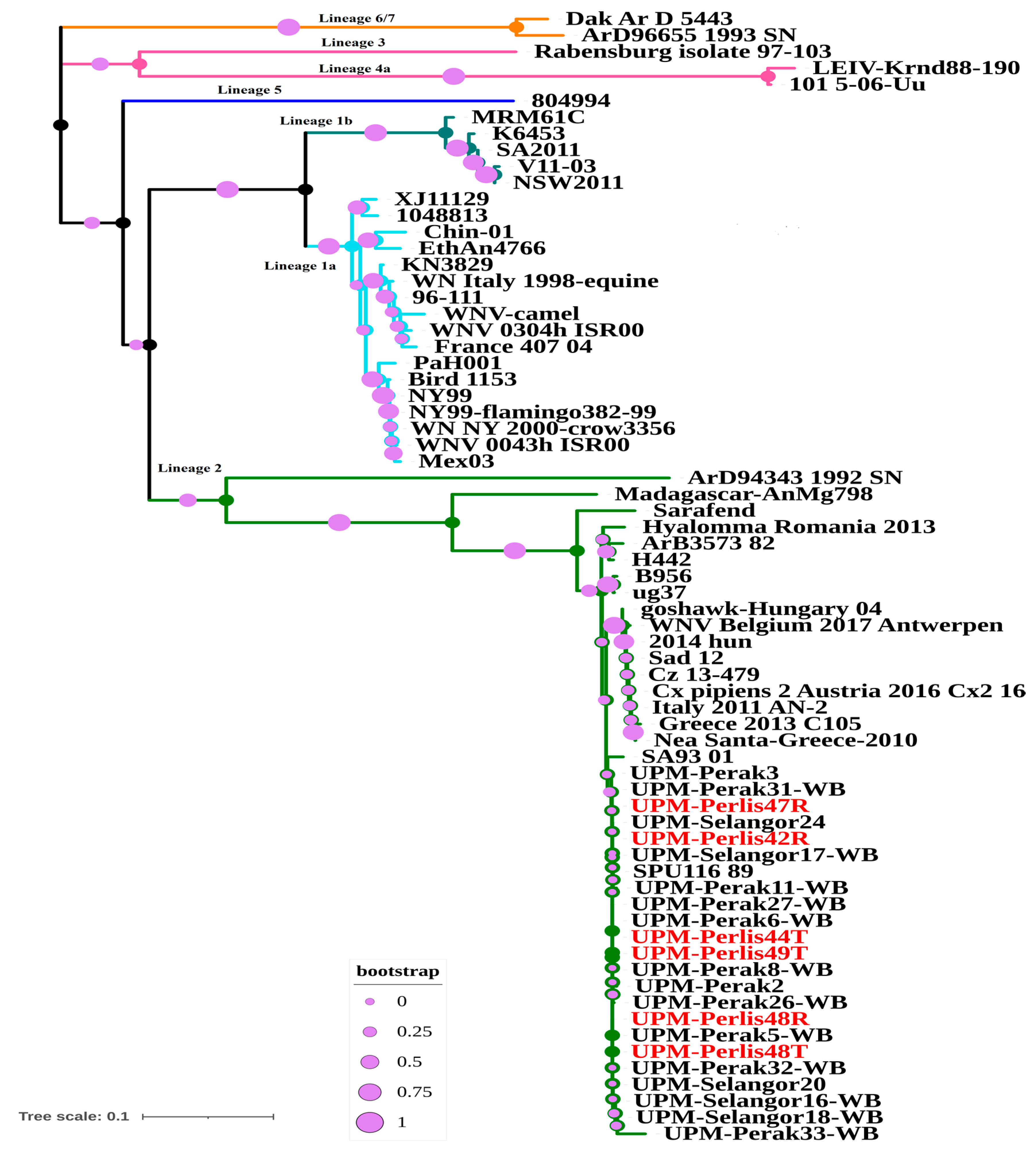
3.2. Molecular Analysis of West Nile Virus in Bats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bunde, J.M.; Heske, E.J.; Mateus-Pinilla, N.E.; Hofmann, J.E.; Novak, R.J. A survey for West Nile virus in bats from Illinois. J. Wildl. Dis. 2006, 42, 455–458. [Google Scholar] [CrossRef]
- Kulkarni, M.A.; Berrang-Ford, L.; Buck, P.A.; Drebot, M.A.; Lindsay, L.R.; Ogden, N.H. Major emerging vector-borne zoonotic diseases of public health importance in Canada. Emerg. Microbes Infect. 2015, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lwande, O.W.; Lutomiah, J.; Obanda, V.; Gakuya, F.; Mutisya, J.; Mulwa, F.; Michuki, G.; Chepkorir, E.; Fischer, A.; Venter, M.; et al. Isolation of tick and mosquito-borne arboviruses from ticks sampled from livestock and wild animal hosts in Ijara District, Kenya. Vector Borne Zoonotic Dis. 2013, 13, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Londono-Renteria, B.; Colpitts, T.M. A Brief Review of West Nile Virus Biology. Methods Mol Biol. 2016, 1435, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chancey, C.; Griney, A.; Volkova, E.; Rios, M. The global ecology and epidemiology of West Nile virus. BioMed Res. Int. 2015, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Root, J.J. West Nile virus associations in wild mammals: A synthesis. Arch. Virol. 2013, 158, 735–752. [Google Scholar] [CrossRef] [PubMed]
- Ølberg, R.A.; Barker, I.K.; Crawshaw, G.J.; Bertelsen, M.F.; Drebot, M.A.; Andonova, M. West Nile virus encephalitis in a Barbary macaque (Macaca sylvanus). Emerg. Infect Dis. 2004, 10, 712–714. [Google Scholar] [CrossRef]
- Ratterree, M.S.; Gutierrez, R.A.; Travassos da Rosa, A.P.; Dille, B.J.; Beasley, D.W.C.; Bohm, R.P.; Desai, S.M.; Didier, P.J.; Bikenmeyer, L.G.; Dawson, G.J.; et al. Experimental infection of rhesus macaques with West Nile virus: Level and duration of viremia and kinetics of the antibody response after infection. J. Infect. Dis. 2004, 189, 669–676. [Google Scholar] [CrossRef]
- Kading, R.C.; Schountz, T. Flavivirus infections of bats: Potential role in Zika virus ecology. Am. J. Trop. Med. Hyg. 2016, 95, 993–996. [Google Scholar] [CrossRef]
- Pilipski, J.D.; Pilipskl, L.M.; Risley, L.S. West nile virus antibodies in bats from New Jersey and New York. J. Wildl. Dis. 2004, 40, 335–337. [Google Scholar] [CrossRef] [PubMed]
- Plowright, R.K.; Parrish, C.R.; McCallum, H.; Hudson, P.J.; Ko, A.I.; Graham, A.L.; Lloyd-Smith, J.O. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017, 15, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Low, V.L.; Chen, C.D.; Lee, H.L.; Lim, P.E.; Leong, C.S.; Sofian-Azirun, M. Nationwide distribution of Culex mosquitoes and associated habitat characteristics at residential areas in Malaysia. J. Am. Mosq. Control Assoc. 2012, 28, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Marlina, S.; Muhd Radzi, S.F.; Lani, R.; Sieng, K.C.; Rahim, N.F.A.; Hassan, H.; Li-Yen, C.; AbuBakar, S.; Zandi, K. Seroprevalence screening for the West Nile virus in Malaysia’s orang asli population. Parasites Vectors 2014, 7, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rais, M.N.; Omar, A.R.; Jalila, A.; Hussni, O.M. Prevalence of West Nile virus antibody in captive bird populations in selected areas in Selangor, Malaysia. In Proceedings of the 6th Seminar on Veterinary Sciences, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Selangor, Malaysia, 11–14 January 2011; Vulume 127. [Google Scholar]
- Ain-Najwa, M.Y.; Yasmin, A.R.; Omar, A.R.; Arshad, S.S.; Abu, J.; Mohammed, H.O.; Kumar, K.; Loong, S.K.; Rovie-Ryan, J.J.; Mohd-Kharip-Shah, A.-K. Evidence of West Nile virus infection in migratory and resident wild birds in West Coast of Peninsular Malaysia. One Health. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Ching, C.Y.; Casals, J.; Bowen, E.T.; Simpson, D.I.; Platt, G.S.; Way, H.J.; Smith, C.E. Arbovirus infections in Sarawak: The isolation of Kunjin virus from mosquitoes of the Culex pseudovishnui group. Ann. Trop. Med. Parasitol. 1970, 64, 263–268. [Google Scholar] [CrossRef]
- Mohammed, M.N.; Yasmin, A.R. West Nile virus: Measures against emergence in Malaysia. J. Vet. Sci. Res. 2019, 4, 1–6. [Google Scholar] [CrossRef]
- Ain-Najwa, M.Y.; Yasmin, A.R.; Omar, A.R.; Arshad, S.S.; Abu, J.; Mohammed, H.O. West Nile Virus Infection in Human and Animals: Potential Risks in Malaysia. Sains Malays. 2019, 48, 2727–2735. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 30–11030. [Google Scholar] [CrossRef]
- Marini, G.; Rosà, R.; Pugliese, A.; Rizzoli, A.; Rizzo, C.; Russo, F.; Montarsi, F.; Capelli, G. West Nile virus transmission and human infection risk in Veneto (Italy): A modelling analysis. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Hii, Y.L.; Zaki, R.A.; Aghamohammadi, N.; Rocklöv, J. Research on climate and dengue in Malaysia: A systematic review. Curr. Environ. Health Rep. 2016, 3, 81–90. [Google Scholar] [CrossRef]
- Di Sabatino, D.; Bruno, R.; Sauro, F.; Danzetta, M.L.; Cito, F.; Ianetti, S.; Narcisi, V.; De Massis, F.; Calistri, P. Epidemiology of West Nile virus disease in Europe and in the Mediterranean Basin from 2019 to 2013. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Murata, R.; Hashiguchi, K.; Yoshii, K.; Kariwa, H.; Nakajima, K.; Ivanov, G.N.; Leonova, G.N.; Takashima, I. Seroprevalence of West Nile virus in wild birds in far eastern Russia using a focus reduction neutralization test. Am. J. Trop. Med. Hyg. 2011, 84, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Mangudo, C.; Aparicio, J.P.; Rossi, G.C.; Gleiser, R.M. Tree hole mosquito species composition and relative abundances differ between urban and adjacent forest habitats in north western Argentina. Bull. Entomol. Res. 2018, 108, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Ratterree, M.S.; Amelia, P.A.; Travassos da Rosa, A.P.A.; Bohm Jr, R.P.; Cogswell, F.B.; Phillippi, K.M.; Caillouet, K.; Schwanberger, S.; Shope, R.E.; Tesh, R.B. West Nile virus infection in nonhuman primate breeding colony, concurrent with the human epidemic, Southern Louisiana. Emerg. Infect. Dis. 2003, 9, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.K.; Kilpatrick, A.M.; Stroud, F.C.; Paul, K.; Wolf, F.; Else, J.G. Seroprevalence of West Nile virus in nonhuman primates as related to mosquito abundance at two national primate research centres. Comp. Med. 2007, 57, 115–119. [Google Scholar]
- Gay, N.; Olival, K.J.; Bumrungsri, S.; Siriaroonrat, B.; Bourgarel, M.; Morand, S. Parasite and viral species richness of Southeast Asian bats: Fragmentation of area distribution matters. Int. J. Parasitol. Parasites Wildl. 2014, 3, 161–170. [Google Scholar] [CrossRef]
- Field, H.; Epstein, J.H.; Henipavirus. Investigating the Role of Bats in Emerging Zoonoses, Balancing Ecology, Conservation and Public Health Interests. In FAO Animal Production and Health Manual; Newman, S., Field, H.E., De Jong, C.E., Epstein, J.H., Eds.; Univerza v Mariboru: Maribor, Slovenia, 2011; Volume 12, p. 64. [Google Scholar]
- Balboni, A.; Battilani, M.; Prosperi, S. The SARS-like coronaviruses: The role of bats and evolutionary relationships with SARS coronavirus. New Microbiol. 2012, 35, 1–16. [Google Scholar]
- Barclay, R.M.R.; Bell, G.P. Marketing and observational techniques. In Ecological and Behavioural Methods for the Study of Bats, 2nd ed.; Smithsonian Institution Press: Washington, DC, USA, 1990; pp. 59–76. [Google Scholar]
- May, F.J.; Davis, C.T.; Tesh, R.B.; Barrett, A.D.T. Phylogeography of West Nile virus: From the cradle of evolution in Africa to Eurasia, Australia, and the Americas. J. Virol. 2011, 85, 2964–2974. [Google Scholar] [CrossRef]
- Venter, M.; Human, S.; Zaayman, D.; Gerdes, G.H.; Williams, J.; Steyl, J.; Leman, P.A.; Paweska, J.T.; Setzkorn, H.; Rous, G.; et al. Lineage 2 West Nile Virus as Cause of Fatal Neurologic Disease in Horses. S. Afr. Emerg. Infect. Dis. 2009, 15, 877–884. [Google Scholar] [CrossRef]
- Botha, E.M.; Markotter, W.; Wolfaardt, M.; Paweska, J.T.; Swanepoel, R.; Palacios, G.; Nel, L.H.; Venter, M. Genetic determinants of virulence in pathogenic lineage 2 West Nile virus strains. Emerg. Infect. Dis. 2008, 14, 222–230. [Google Scholar] [CrossRef]
- Petruccelli, A.; Zottola, T.; Ferrara, G.; Iovane, V.; Russo, C.D.; Pagnini, U.; Montagnaro, S. West Nile virus and related flavivirus in European wild boar (Suc scrofa), Latium region, Italy: A retrospective study. Animals 2020, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.B.F.; Göertz, G.P.; Pjilman, G.P.; Koenraadt, C.J. Vector competence of European mosquitoes for West Nile virus. Emerg. Microbes Infect. 2017, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, J.; Chirico, J.; Boqvist, S.; Thu, H.T.V.; Magnusson, U. Occurrence of Japanese Encephalitis virus mosquito vectors in relation to urban pig holdings. Am. J. Trop. Med. Hyg. 2012, 87, 1076–1082. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.L.; Anderson, S.L.; Smartt, C.T. Vector competence of Florida mosquitoes for chikungunya virus. J. Vector Ecol. 2010, 35, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Marini, G.; Poletti, P.; Giacobini, M.; Pugliese, A.; Merler, S.; Rosà, R. The role of climatic and density-dependent factors in shaping mosquito population dynamics: The case of Culex pipiens in Northwestern Italy. PLoS ONE 2016, 11, e0154018. [Google Scholar] [CrossRef] [PubMed]
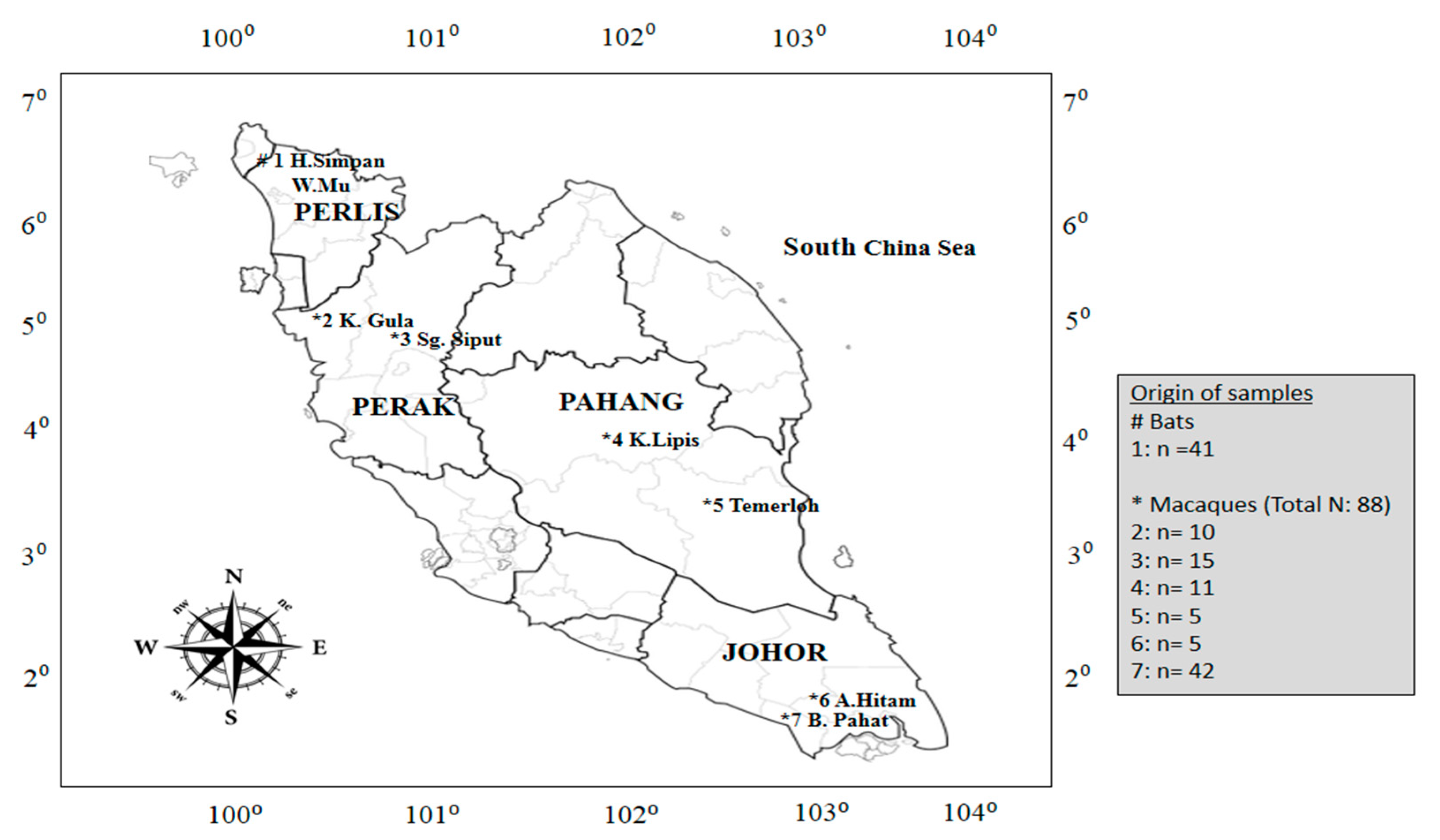
| Family | Species | Malaysian State | No. of Macaques | No. of Serum Sample | No. of Positive WNV Antibody (c-ELISA) | No. of Oropharyngeal Swabs | No. of Positive WNV RNA (RT-PCR) |
|---|---|---|---|---|---|---|---|
| Cercopithecidae | Long-tailed macaque (Macaca Fascicularis) | Pahang | 8 | 6 | 1 | 8 | 0 |
| 0 | 0 | - | 0 | - | |||
| 4 | 2 | 1 | 4 | 0 | |||
| 0 | 0 | - | 0 | - | |||
| 0 | 0 | - | 0 | - | |||
| 4 | 1 | 0 | 3 | 0 | |||
| Perak | 5 | 5 | 3 | 0 | - | ||
| 0 | 0 | - | 0 | - | |||
| 11 | 11 | 2 | 1 | 0 | |||
| 5 | 5 | 2 | 0 | - | |||
| 0 | 0 | - | 0 | - | |||
| 4 | 4 | 1 | 0 | 0 | |||
| Johor | 5 | 5 | 1 | 5 | 0 | ||
| 3 | 3 | 0 | 3 | 0 | |||
| 21 | 21 | 8 | 21 | 0 | |||
| 6 | 6 | 0 | 6 | 0 | |||
| 2 | 2 | 0 | 2 | 0 | |||
| 10 | 10 | 5 | 10 | 0 | |||
| Total | 88 | 81 | 24 | 63 | 0 | ||
| Family | Species | No. of Bats | No. of Oropharyngeal Swabs | No. of Rectal Swabs | No. of Positive WNV RNA (RT-PCR) |
|---|---|---|---|---|---|
| Pteropodidae | Horsfield’s Fruit Bats (Cynopterus horsfieldii) | 1 | 1 | 1 | 0 |
| 5 | 4 | 5 | 0 | ||
| 2 | 2 | 2 | 0 | ||
| 1 | 1 | 1 | 0 | ||
| Lesser Short-nosed Fruit Bats (Cynopterus brachyotis) | 0 | 0 | 0 | - | |
| 0 | 0 | 0 | - | ||
| 1 | 1 | 1 | 1 | ||
| 0 | 0 | 0 | - | ||
| Long-tongued Fruit Bats (Macroglossus sobrinus) | 0 | 0 | 0 | - | |
| 1 | 1 | 1 | 0 | ||
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| Emballonuridae | Lesser Sheath-tailed Bats (Emballonura monticola) | 0 | 0 | 0 | - |
| 3 | 3 | 3 | 2 | ||
| 1 | 1 | 1 | 0 | ||
| 2 | 2 | 2 | 1 | ||
| Rhinolophidae | Blyth’s Horseshoe Bats (Rhinolophus lepidus) | 1 | 1 | 1 | 0 |
| 2 | 1 | 2 | 0 | ||
| 5 | 1 | 5 | 0 | ||
| 2 | 1 | 2 | 0 | ||
| Malayan Horseshoe Bats (Rhinolophus malayanus) | 0 | 0 | 0 | 0 | |
| 2 | 2 | 2 | 0 | ||
| 0 | 0 | 0 | 0 | ||
| 0 | 0 | 0 | 0 | ||
| Thai Horseshoe Bats (Rhinolophus siamensis) | 1 | 1 | 1 | 2 | |
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| Croslet Horseshoe Bats (Rhinolophus coelophyllus) | 0 | 0 | 0 | - | |
| 1 | 1 | 1 | 0 | ||
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| Bourret’s Horseshoe Bats (Rhinolophus paradoxolophus) | 0 | 0 | 0 | - | |
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| 1 | 1 | 1 | 0 | ||
| Hipposideridae | Great Roundleaf Bats (Hipposideros armiger) | 1 | 1 | 1 | 0 |
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| 1 | 1 | 1 | 0 | ||
| Shield-faced Roundleaf Bats (Hipposideros lylei) | 1 | 1 | 0 | 0 | |
| 1 | 1 | 1 | 0 | ||
| 2 | 2 | 1 | 0 | ||
| 1 | 1 | 0 | 0 | ||
| Diadem Leaf-nosed Bats (Hipposideros diadema) | 0 | 0 | 0 | - | |
| 1 | 1 | 1 | 0 | ||
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| Vespertilionidae | Lesser Asiatic Yellow Bats (Scotophilus kuhlii) | 0 | 0 | 0 | - |
| 1 | 1 | 1 | 0 | ||
| 0 | 0 | 0 | - | ||
| 0 | 0 | 0 | - | ||
| TOTAL | 41 | 34 | 38 | Bats +ve: 5 RT-PCR +ve: 6 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ain-Najwa, M.Y.; Yasmin, A.R.; Arshad, S.S.; Omar, A.R.; Abu, J.; Kumar, K.; Mohammed, H.O.; Natasha, J.A.; Mohammed, M.N.; Bande, F.; et al. Exposure to Zoonotic West Nile Virus in Long-Tailed Macaques and Bats in Peninsular Malaysia. Animals 2020, 10, 2367. https://doi.org/10.3390/ani10122367
Ain-Najwa MY, Yasmin AR, Arshad SS, Omar AR, Abu J, Kumar K, Mohammed HO, Natasha JA, Mohammed MN, Bande F, et al. Exposure to Zoonotic West Nile Virus in Long-Tailed Macaques and Bats in Peninsular Malaysia. Animals. 2020; 10(12):2367. https://doi.org/10.3390/ani10122367
Chicago/Turabian StyleAin-Najwa, Mohd Yuseri, Abd Rahaman Yasmin, Siti Suri Arshad, Abdul Rahman Omar, Jalila Abu, Kiven Kumar, Hussni Omar Mohammed, Jafar Ali Natasha, Mohammed Nma Mohammed, Faruku Bande, and et al. 2020. "Exposure to Zoonotic West Nile Virus in Long-Tailed Macaques and Bats in Peninsular Malaysia" Animals 10, no. 12: 2367. https://doi.org/10.3390/ani10122367
APA StyleAin-Najwa, M. Y., Yasmin, A. R., Arshad, S. S., Omar, A. R., Abu, J., Kumar, K., Mohammed, H. O., Natasha, J. A., Mohammed, M. N., Bande, F., Abdullah, M.-L., & J. Rovie-Ryan, J. (2020). Exposure to Zoonotic West Nile Virus in Long-Tailed Macaques and Bats in Peninsular Malaysia. Animals, 10(12), 2367. https://doi.org/10.3390/ani10122367






