Genomic Characterization of the Istrian Shorthaired Hound
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. SNP Genotyping, Assembly of the Merged Dataset, and Quality Control
2.3. Genealogical Analysis
2.4. Genetic Parameters
2.5. Runs of Homozygosity
2.6. Genetic Relationships and Population Structure
2.6.1. Genomic Relationships and Genetic Distances
2.6.2. Population Structure
2.7. Genome-Wide Scan of Selection Signatures
3. Results
3.1. Genealogical Data
3.2. Population Genetics Statistics
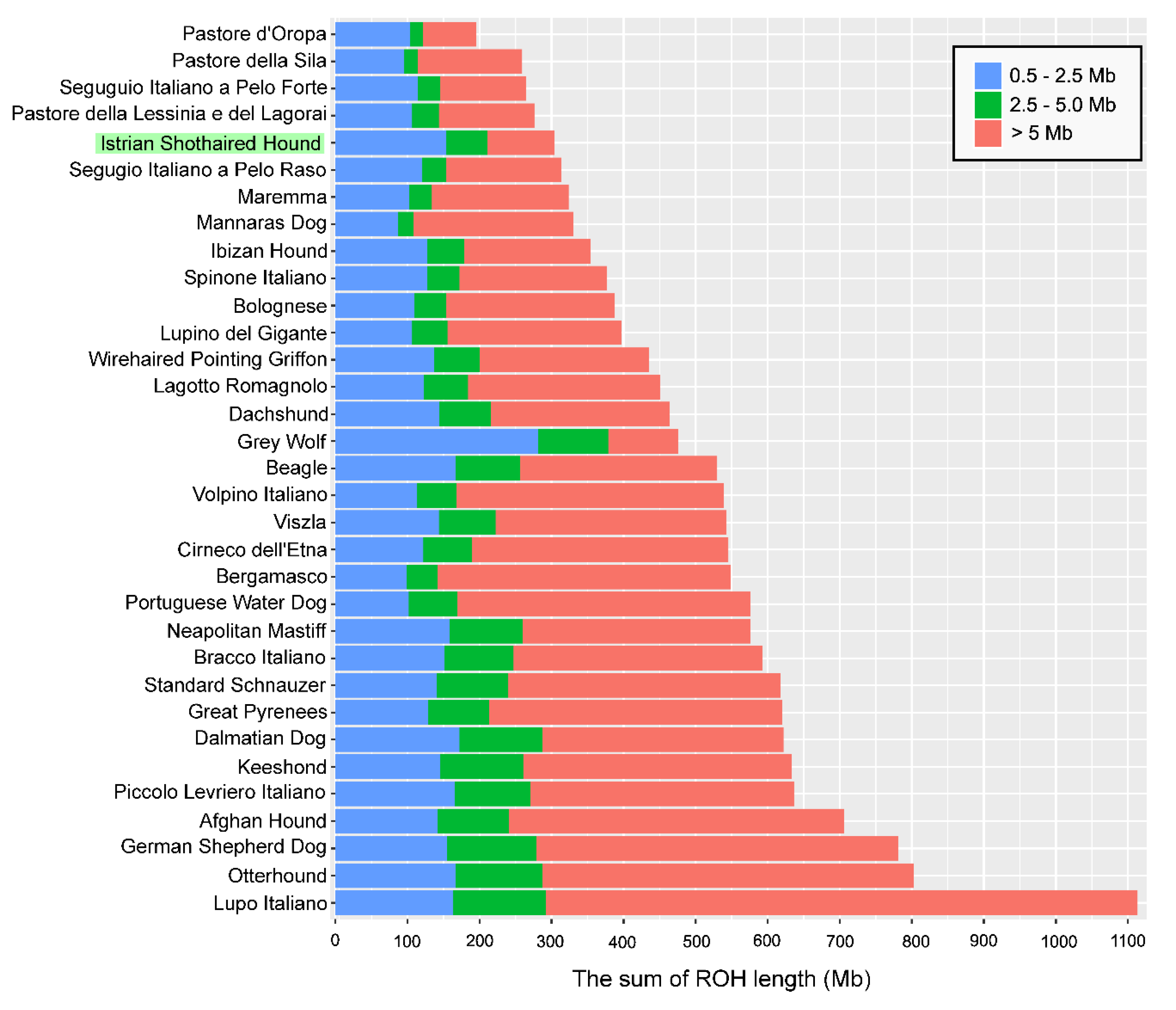
3.3. Runs of Homozygosity
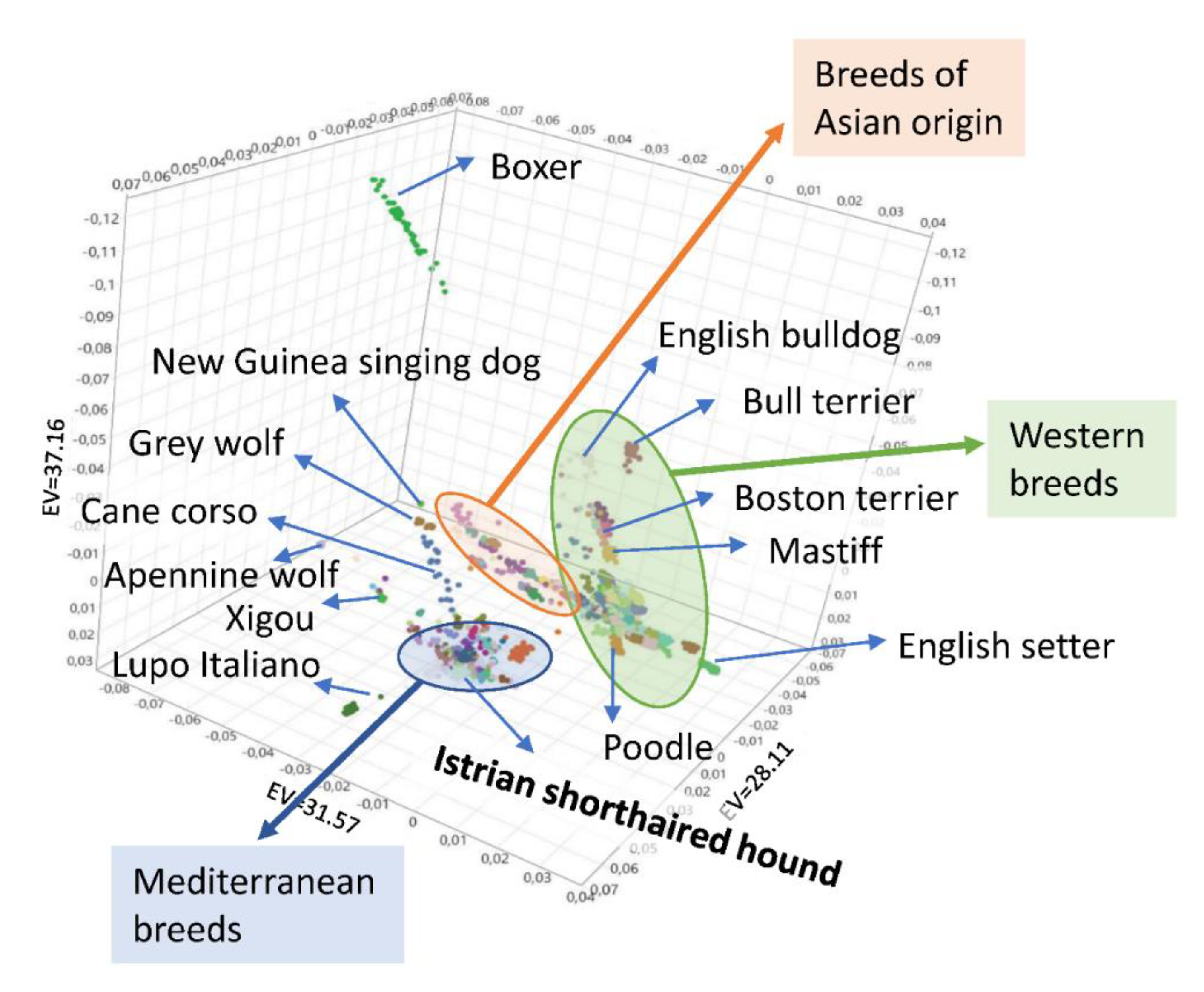
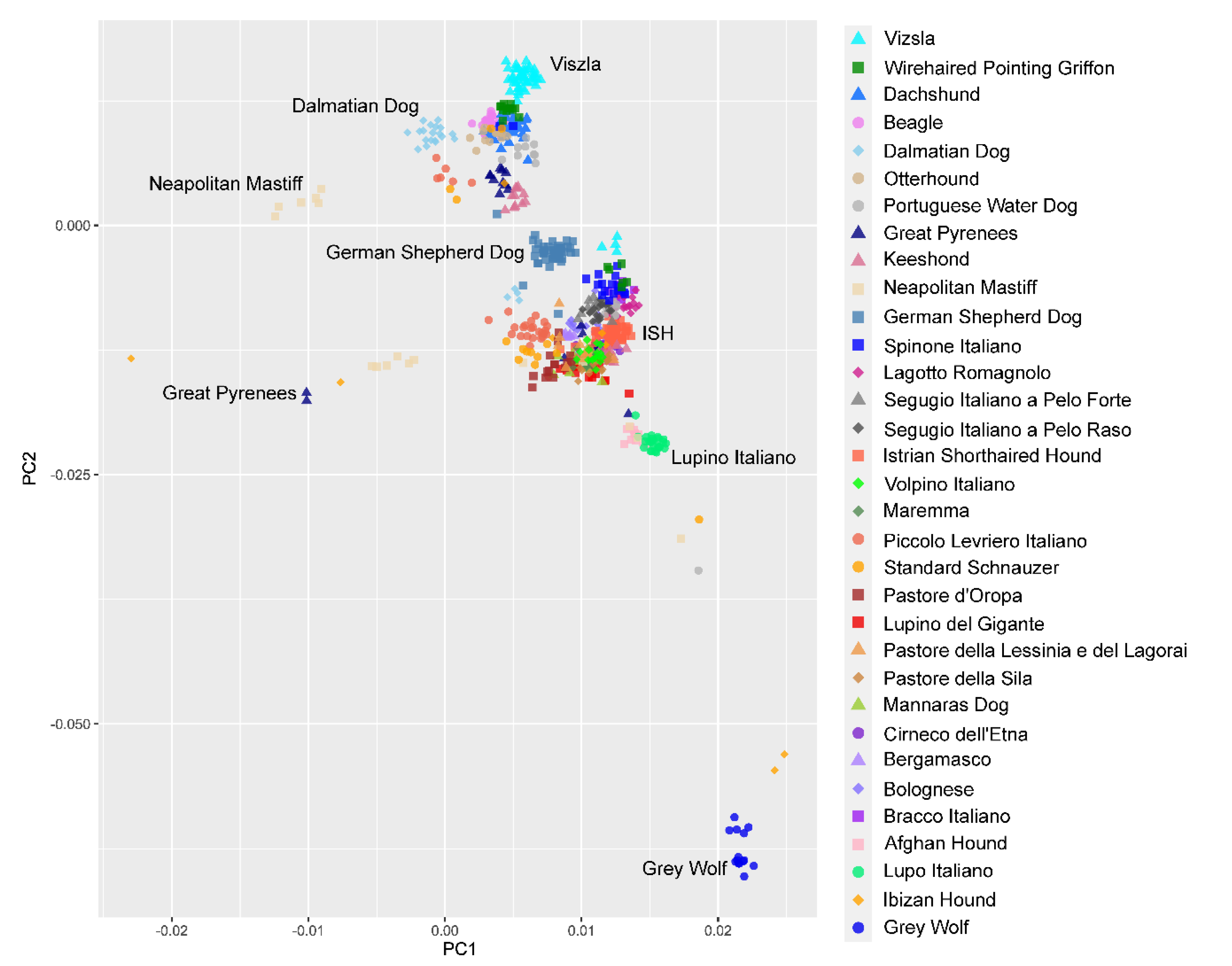
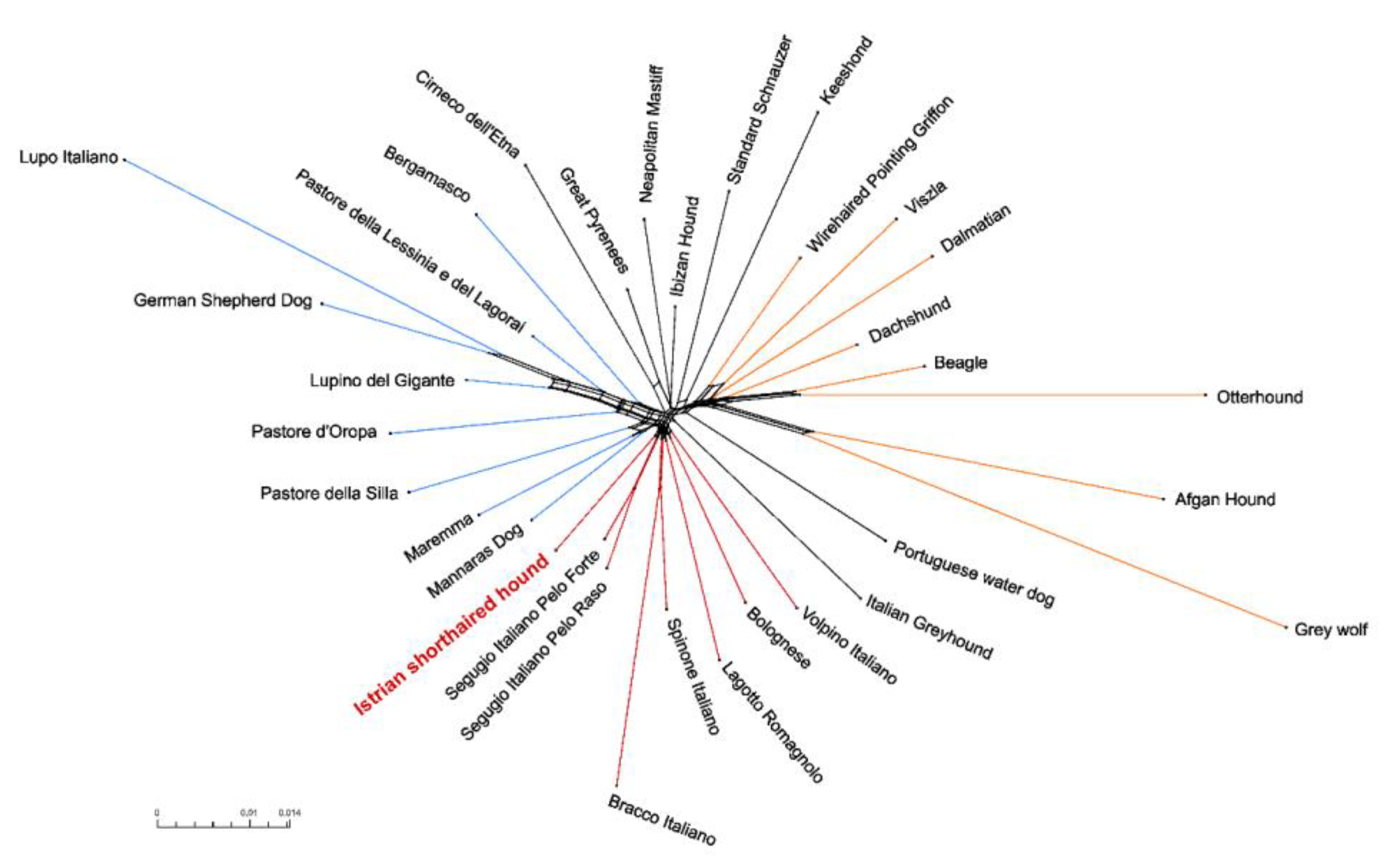
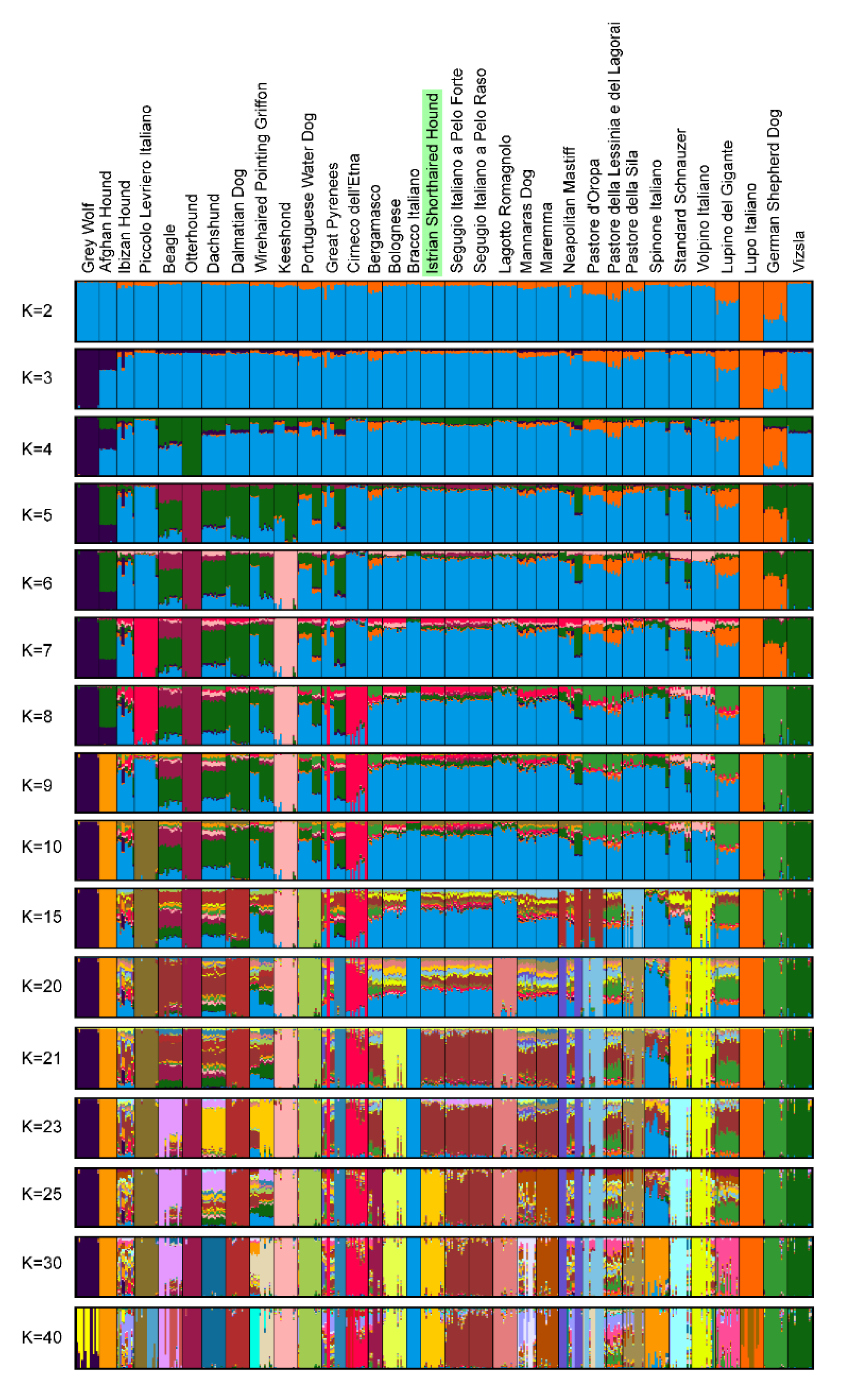
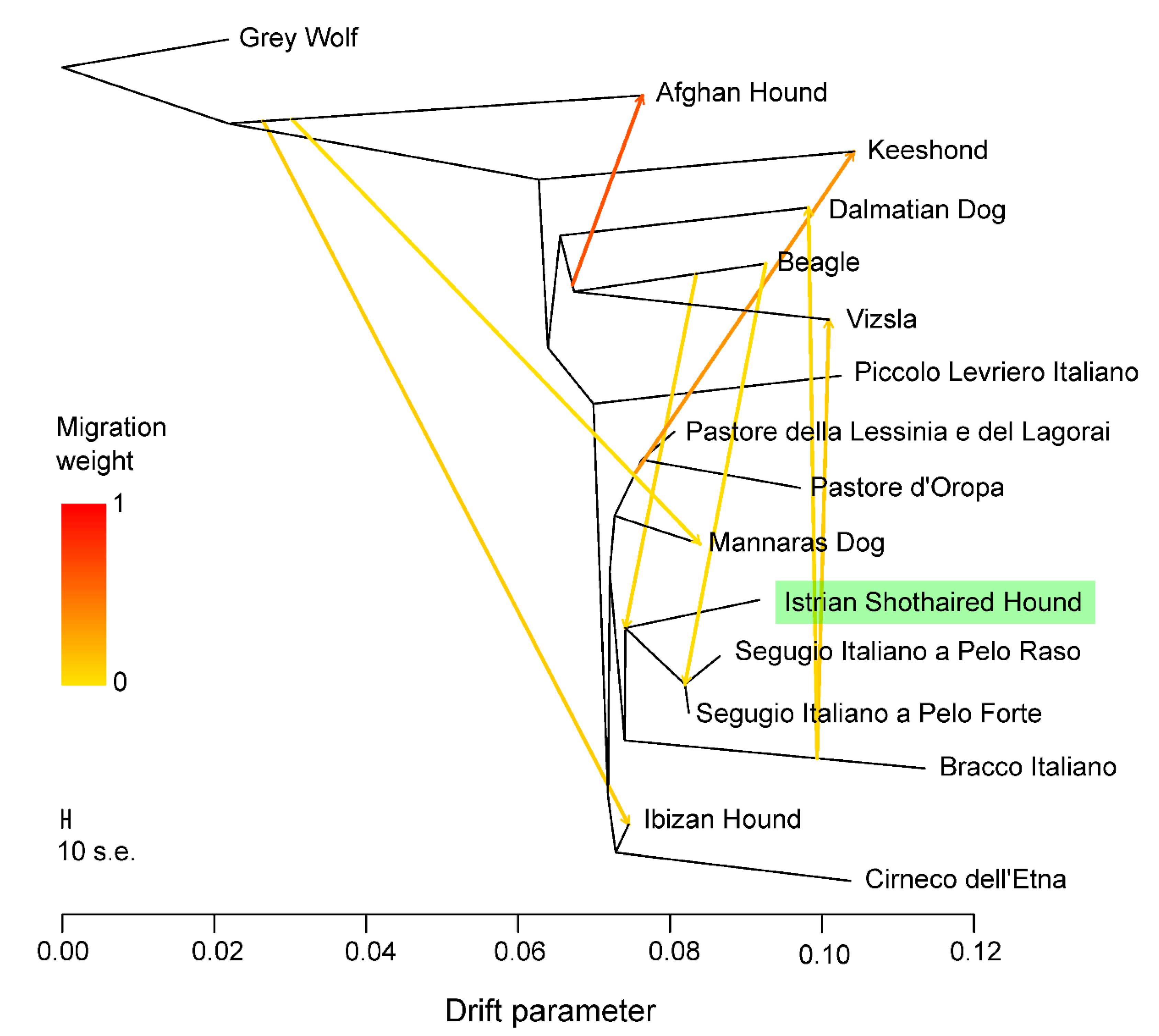
3.4. Genetic Relationships and Population Structure
3.5. Genomic Patterns of Homozygosity
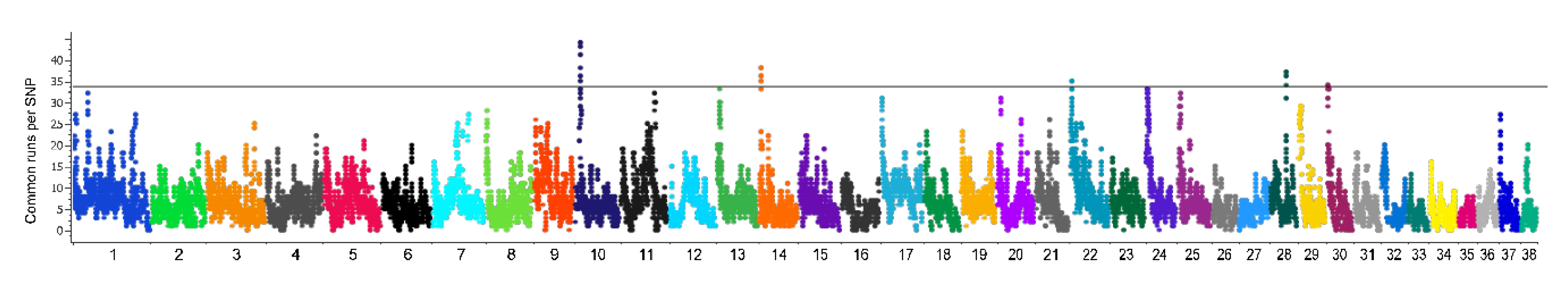
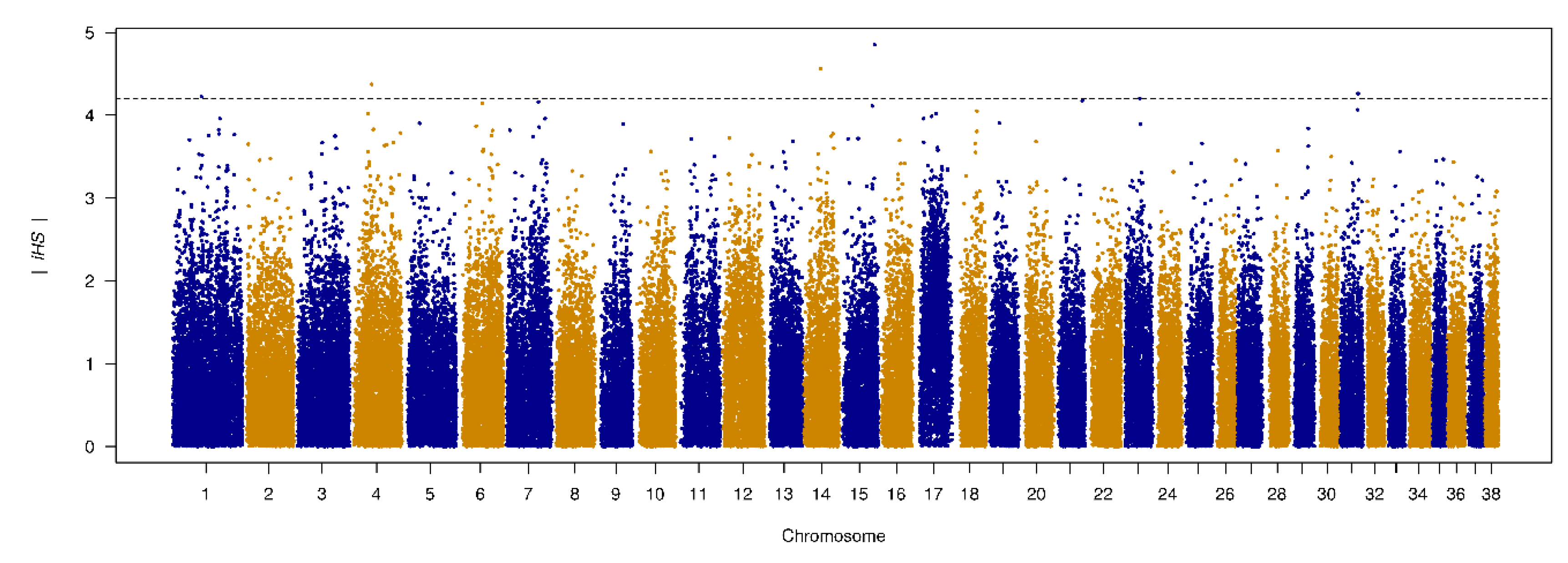
3.6. Genome-Wide Scan of Genome Signatures
4. Discussion
4.1. Population Parameters, Inbreeding, and Effective Population Size
4.2. Clustering of Breeds
4.3. Phenotypic and Hunting Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Romić, S. Uz 470. godišnjicu ergele đakovačke. Zb. Muz. Đakovštine 1978, 1, 27–39. [Google Scholar]
- Croatian Kennel Club. Croatian Kennel Club—Ilica 61, HR—10000 Zagreb. Available online: http://webeng.hks.hr/ (accessed on 9 October 2020).
- Pasarić, O. Breeding Characteristics of Croatian Hunting Dog Breeds on the Territory of Croatia. Master’s Thesis, Josip Juraj Strossmayer University of Osijek, Osijek, Croatia, 2018. [Google Scholar]
- Broeckx, B.J.G. The dog 2.0: Lessons learned from the past. Theriogenology 2020, 150, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Zajc, I.; Kus, M. Populacijska struktura dalmatincev in istrskih gonicev ter njihovi evolucijski odnosi z nekaterimi drugimi pasmami, kot jih nakazujejo polimorfni mikrosateliti = Population structure of Dalmatians and Istrian shorthaired hounds and their evolutionary relationships to some other breeds as revealed by polymorhic microsatellites. Slov. Vet. Res. 2001, 38, 35–43. [Google Scholar]
- Zajc, I.; Mellersh, C.S.; Sampson, J. Variability of canine microsatellites within and between different dog breeds. Mamm. Genome 1997, 8, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Chen, L.; Wang, S.; Oh, C.; Zhao, H. Comparison of single-nucleotide polymorphisms and microsatellites in inference of population structure. BMC Genet. 2005, 6, S26. [Google Scholar] [CrossRef] [PubMed]
- Dreger, D.L.; Davis, B.W.; Cocco, R.; Sechi, S.; Di Cerbo, A.; Parker, H.G.; Polli, M.; Marelli, S.P.; Crepaldi, P.; Ostrander, E.A. Commonalities in Development of Pure Breeds and Population Isolates Revealed in the Genome of the Sardinian Fonni’s Dog. Genetics 2016, 204, 737–755. [Google Scholar] [CrossRef] [PubMed]
- Talenti, A.; Dreger, D.L.; Frattini, S.; Polli, M.; Marelli, S.; Harris, A.C.; Liotta, L.; Cocco, R.; Hogan, A.N.; Bigi, D.; et al. Studies of modern Italian dog populations reveal multiple patterns for domestic breed evolution. Ecol. Evol. 2018, 8, 2911–2925. [Google Scholar] [CrossRef]
- Parker, H.G.; Dreger, D.L.; Rimbault, M.; Davis, B.W.; Mullen, A.B.; Carpintero-Ramirez, G.; Ostrander, E.A. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridisation on Modern Dog Breed Development. Cell Rep. 2017, 19, 697–708. [Google Scholar] [CrossRef]
- Shannon, L.M.; Boyko, R.H.; Castelhano, M.; Corey, E.; Hayward, J.J.; McLean, C.; White, M.E.; Abi Said, M.; Anita, B.A.; Bondjengo, N.I.; et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc. Natl. Acad. Sci. USA 2015, 112, 13639–13644. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Iwaisaki, H.; Colleau, J.J. CFC: A tool for monitoring genetic diversity. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Minas Gerais, Brazil, 13–18 August 2006. [Google Scholar]
- Gutierrez, J.P.; Altarriba, J.; Diaz, C.; Quintanilla, R.; Canon, J.; Piedrafita, J. Pedigree analysis of eight Spanish beef cattle breeds. Genet. Sel. Evol. 2003, 35, 43–63. [Google Scholar] [CrossRef]
- Gutierrez, J.P.; Goyache, F. A note on ENDOG: A computer program for analysing pedigree information. J. Anim. Breed. Genet. 2005, 122, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S. Introduction to Quantitative Genetics; Prentice Hall: Harlow, UK, 1996. [Google Scholar]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- McQuillan, R.; Leutenegger, A.-L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Cogan, N.O.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localised haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef]
- R Consortium Team. R: The R Project for Statistical Computing. 2020. Available online: https://www.R-project.org/ (accessed on 31 October 2020).
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef]
- Ceballos, F.C.; Joshi, P.K.; Clark, D.W.; Ramsay, M.; Wilson, J.F. Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 2018, 19, 220–234. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, Y.; Wang, L.; Liu, X.; Yan, H.; Zhang, L. Molecular cloning of WIF1 and HMGA2 reveals ear-preferential expression while uncovering a missense mutation associated with porcine ear size in WIF1. Anim. Genet. 2019, 50, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Plassais, J.; Kim, J.; Davis, B.W.; Karyadi, D.M.; Hogan, A.N.; Harris, A.C.; Decker, B.; Parker, H.G.; Ostrander, E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Laramie, J.M.; Wilk, J.B.; Williamson, S.L.; Nagle, M.W.; Latourelle, J.C.; Tobin, J.E.; Province, M.A.; Borecki, I.B.; Myers, R.H. Polymorphisms near EXOC4 and LRGUK on chromosome 7q32 are associated with Type 2 Diabetes and fasting glucose; the NHLBI Family Heart Study. BMC Med. Genet. 2008, 9, 46. [Google Scholar] [CrossRef]
- Akkad, D.A.; Gerding, W.M.; Gasser, R.B.; Epplen, J.T. Homozygosity mapping and sequencing identify two genes that might contribute to pointing behavior in hunting dogs. Canine Genet. Epidemiol. 2015, 2, 5. [Google Scholar] [CrossRef]
- Kim, J.; Williams, F.J.; Dreger, D.L.; Plassais, J.; Davis, B.W.; Parker, H.G.; Ostrander, E.A. Genetic selection of athletic success in sport-hunting dogs. Proc. Natl. Acad. Sci. USA 2018, 115, E7212. [Google Scholar] [CrossRef]
- Cecchi, F.; Carlini, G.; Giuliotti, L.; Russo, C. Inbreeding may affect phenotypic traits in an Italian population of Basset Hound dogs. Rend. Lincei. Sci. Fis. Nat. 2018, 29, 165–170. [Google Scholar] [CrossRef]
- Kettunen, A.; Daverdin, M.; Helfjord, T.; Berg, P. Cross-Breeding Is Inevitable to Conserve the Highly Inbred Population of Puffin Hunter: The Norwegian Lundehund. PLoS ONE 2017, 12, e0170039. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, E.; Slawinska, A.; Mroczkowski, S. Analiza poziomu inbredu w polskiej populacji psów rasy alpejski gończy krótkonożny w latach 2000–2016. Rocz. Nauk. Pol. Tow. Zootech. 2018, 14, 9–18. [Google Scholar]
- Karjalainen, L.; Ojala, M. Generation intervals and inbreeding coefficients in the Finnish Hound and the Finnish Spitz. J. Anim. Breed. Genet. 1997, 114, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Voges, S.; Distl, O. Inbreeding trends and pedigree analysis of Bavarian mountain hounds, Hanoverian hounds and Tyrolean hounds. J. Anim. Breed. Genet. 2009, 126, 357–365. [Google Scholar] [CrossRef]
- Mäki, K.; Groen, A.F.; Liinamo, A.E.; Ojala, M. Population structure, inbreeding trend and their association with hip and elbow dysplasia in dogs. Anim. Sci. 2001, 73, 217–228. [Google Scholar] [CrossRef]
- Cecchi, F.; Paci, G.; Spaterna, A.; Ragatzu, M.; Ciampolini, R. Demographic approach on the study of genetic parameters in the dog Braque Français type Pyrénées Italian population. Ital. J. Anim. Sci. 2016, 15, 30–36. [Google Scholar] [CrossRef]
- Gresky, C.; Hamann, H.; Distl, O. Einfluss von Inzucht auf die Wurfgröße und den Anteil tot geborener Welpen beim Dackel. Berl. Und Münchener Tierärztliche Wochenschr. 2005, 118, 134–139. [Google Scholar]
- Goleman, M.; Balicki, I.; Radko, A.; Jakubczak, A.; Fornal, A. Genetic diversity of the Polish Hunting Dog population based on pedigree analyses and molecular studies. Livest. Sci. 2019, 229, 114–117. [Google Scholar] [CrossRef]
- Mortlock, S.A.; Khatkar, M.S.; Williamson, P. Comparative Analysis of Genome Diversity in Bullmastiff Dogs. PLoS ONE 2016, 11, e0147941. [Google Scholar] [CrossRef]
- Boichard, D.; Maignel, L.; Verrier, É. The value of using probabilities of gene origin to measure genetic variability in a population. Genet. Sel. Evol. 1997, 29, 5. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G.; Mary-Huard, T.; Verrier, E.; Danvy, S.; Charvolin, E.; Danchin-Burge, C. Methods to estimate effective population size using pedigree data: Examples in dog, sheep, cattle and horse. Genet. Sel. Evol. 2013, 45, 1. [Google Scholar] [CrossRef] [PubMed]
- Curik, I.; Ferencakovic, M.; Solkner, J. Genomic dissection of inbreeding depression: A gate to new opportunities. Rev. Bras. De Zootec. Braz. J. Anim. Sci. 2017, 46, 773–782. [Google Scholar] [CrossRef]
- Nicholas, F.W.; Wade, C.M.; Williamson, P. Disorders in pedigree dogs: Assembling the evidence. Vet. J. 2010, 183, 8–9. [Google Scholar] [CrossRef]
- Bauer, M.; Stubičan, Đ.; Labura, Č. Ist der Dalmatiner die einzige Hunderasse mit spezifischem Stoffwechsel der Purinbasen? Wien. Tierarztl. Mon. 1999, 86, 136–139. [Google Scholar]
- Bauer, M.; Lemo, N. The origin and evolution of Dalmatian and relation with other croatian native breeds of dog. Rev. Med. Vet. 2008, 159, 38. [Google Scholar]
- Mastrangelo, S.; Biscarini, F.; Tolone, M.; Auzino, B.; Ragatzu, M.; Spaterna, A.; Ciampolini, R. Genomic characterisation of the Braque Francais type Pyrenees dog and relationship with other breeds. PLoS ONE 2018, 13, e0208548. [Google Scholar] [CrossRef]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef]
- Lippi, G.; Montagnana, M.; Salvagno, G.L.; Franchini, M.; Guidi, G.C. Glycaemic control in athletes. Int. J. Sports Med. 2008, 29, 7–10. [Google Scholar] [CrossRef]
- Freedman, A.H.; Schweizer, R.M.; Ortega-Del Vecchyo, D.; Han, E.; Davis, B.W.; Gronau, I.; Silva, P.M.; Galaverni, M.; Fan, Z.; Marx, P.; et al. Demographically-Based Evaluation of Genomic Regions under Selection in Domestic Dogs. PLoS Genet. 2016, 12, e1005851. [Google Scholar] [CrossRef] [PubMed]
- Ishisaka, M.; Hara, H. The roles of diacylglycerol kinases in the central nervous system: Review of genetic studies in mice. J. Pharmacol. Sci. 2014, 124, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Shirai, Y.; Kouzuki, T.; Kakefuda, K.; Moriguchi, S.; Oyagi, A.; Horie, K.; Morita, S.-y.; Shimazawa, M.; Fukunaga, K.; Takeda, J.; et al. Essential role of neuron-enriched diacylglycerol kinase (DGK), DGKbeta in neurite spine formation, contributing to cognitive function. PLoS ONE 2010, 5, e11602. [Google Scholar] [CrossRef]
- Lee, S.E.; Simons, S.B.; Heldt, S.A.; Zhao, M.; Schroeder, J.P.; Vellano, C.P.; Cowan, D.P.; Ramineni, S.; Yates, C.K.; Feng, Y.; et al. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci USA 2010, 107, 16994–16998. [Google Scholar] [CrossRef] [PubMed]
- López-Aranda, M.F.; López-Téllez, J.F.; Navarro-Lobato, I.; Masmudi-Martín, M.; Gutiérrez, A.; Khan, Z.U. Role of layer 6 of V2 visual cortex in object-recognition memory. Science 2009, 325, 87–89. [Google Scholar] [CrossRef]
- Tejeda, G.S.; Whiteley, E.L.; Deeb, T.Z.; Bürli, R.W.; Moss, S.J.; Sheridan, E.; Brandon, N.J.; Baillie, G.S. Chorea-related mutations in PDE10A result in aberrant compartmentalisation and functionality of the enzyme. Proc. Natl. Acad. Sci. USA 2020, 117, 677. [Google Scholar] [CrossRef]


| Depository | Accession | Number of Samples | Number of Breeds | Reference |
|---|---|---|---|---|
| GEO | GSE121027 | 273 | 21 | [9] |
| GEO | GSE83160 | 137 | 22 | [8] |
| GEO | GSE96736 | 158 | 47 | [10] |
| Dryad | dryad.v9t5h | 5406 | 163 | [11] |
| Dryad | dryad.1vhhmgqrj | 48 | 1 | this study |
| Parameter | Reference Population |
|---|---|
| Number of analyzed animals | 520 |
| Number of inbreeds | 54 |
| Average inbreeding coefficient | 0.42% |
| Average inbreeding coefficient in inbreeds | 4.08% |
| Average relatedness | 1.46% |
| Inbreeding rate per generation, ΔF | 0.45% |
| Effective population size | 111.24 |
| Number of founder animals | 179 |
| The effective number of founders | 100 |
| The effective number of ancestors | 59 |
| Number of maximum generations, mean | 2.26 |
| Number of complete generations, mean | 1.36 |
| Number of equivalent generations, mean | 1.70 |
| Breed | Pol. Loci [%] | N | P | HOBS ± SD | HEXP ± SD | π ± SD | Fis ± SD | FROH ± SD | Ne |
|---|---|---|---|---|---|---|---|---|---|
| Afghan Hound | 70.38 | 10.99 | 0.82 | 0.238 ± 0.214 | 0.237 ± 0.193 | 0.248 ± 0.202 | 0.024 ± 0.233 | 0.295 ± 0.074 | 32 |
| Beagle | 89.87 | 30.97 | 0.78 | 0.277 ± 0.174 | 0.295 ± 0.174 | 0.300 ± 0.176 | 0.062 ± 0.185 | 0.221 ± 0.083 | 88 |
| Bergamasco | 81.04 | 9.00 | 0.79 | 0.276 ± 0.213 | 0.282 ± 0.182 | 0.299 ± 0.193 | 0.050 ± 0.288 | 0.229 ± 0.136 | 28 |
| Bolognese | 88.93 | 17.98 | 0.77 | 0.302 ± 0.191 | 0.304 ± 0.172 | 0.313 ± 0.177 | 0.028 ± 0.220 | 0.162 ± 0.059 | 55 |
| Bracco Italiano | 72.61 | 8.99 | 0.81 | 0.268 ± 0.229 | 0.253 ± 0.193 | 0.268 ± 0.204 | −0.001 ± 0.238 | 0.248 ± 0.025 | 30 |
| Cirneco dell’Etna | 83.93 | 13.87 | 0.80 | 0.275 ± 0.202 | 0.276 ± 0.180 | 0.286 ± 0.187 | 0.027 ± 0.238 | 0.217 ± 0.119 | 38 |
| Dachshund | 93.34 | 39.96 | 0.77 | 0.289 ± 0.166 | 0.310 ± 0.167 | 0.314 ± 0.169 | 0.068 ± 0.167 | 0.194 ± 0.091 | 140 |
| Dalmatian Dog | 84.90 | 22.99 | 0.79 | 0.265 ± 0.188 | 0.282 ± 0.181 | 0.289 ± 0.185 | 0.064 ± 0.220 | 0.260 ± 0.053 | 69 |
| German Shepherd Dog | 89.46 | 47.93 | 0.81 | 0.238 ± 0.174 | 0.255 ± 0.180 | 0.258 ± 0.182 | 0.067 ± 0.167 | 0.326 ± 0.079 | 86 |
| Grey Wolf | 77.20 | 13.53 | 0.82 | 0.200 ± 0.174 | 0.248 ± 0.186 | 0.257 ± 0.193 | 0.158 ± 0.288 | 0.181 ± 0.170 | 66 |
| Great Pyrenees | 94.38 | 19.92 | 0.76 | 0.261 ± 0.157 | 0.322 ± 0.160 | 0.331 ± 0.165 | 0.177 ± 0.265 | 0.259 ± 0.129 | 70 |
| Ibizan Hound | 93.54 | 10.93 | 0.75 | 0.307 ± 0.179 | 0.336 ± 0.166 | 0.352 ± 0.168 | 0.111 ± 0.144 | 0.135 ± 0.107 | 53 |
| Istrian Shothaired Hound | 93.99 | 47.37 | 0.77 | 0.317 ± 0.182 | 0.311 ± 0.154 | 0.314 ± 0.162 | −0.006 ± 0.298 | 0.123 ± 0.033 | 109 |
| Keeshond | 78.60 | 20.98 | 0.80 | 0.259 ± 0.204 | 0.266 ± 0.191 | 0.272 ± 0.196 | 0.032 ± 0.218 | 0.264 ± 0.049 | 50 |
| Lagotto Romagnolo | 86.86 | 17.94 | 0.79 | 0.293 ± 0.198 | 0.287 ± 0.177 | 0.296 ± 0.182 | 0.007 ± 0.205 | 0.188 ± 0.050 | 51 |
| Lupino del Gigante | 89.17 | 15.99 | 0.77 | 0.302 ± 0.192 | 0.303 ± 0.171 | 0.312 ± 0.176 | 0.028 ± 0.224 | 0.166 ± 0.089 | 53 |
| Lupo Italiano | 52.06 | 23.87 | 0.87 | 0.185 ± 0.218 | 0.174 ± 0.198 | 0.178 ± 0.202 | −0.016 ± 0.139 | 0.451 ± 0.037 | 20 |
| Mannaras Dog | 89.94 | 11.99 | 0.76 | 0.314 ± 0.200 | 0.315 ± 0.166 | 0.329 ± 0.173 | 0.040 ± 0.272 | 0.129 ± 0.075 | 42 |
| Maremma | 87.19 | 13.98 | 0.77 | 0.310 ± 0.201 | 0.302 ± 0.173 | 0.313 ± 0.180 | 0.009 ± 0.230 | 0.135 ± 0.056 | 46 |
| Neapolitan Mastiff | 92.05 | 17.95 | 0.77 | 0.271 ± 0.177 | 0.309 ± 0.167 | 0.317 ± 0.172 | 0.125 ± 0.271 | 0.241 ± 0.104 | 60 |
| Otterhound | 62.81 | 11.99 | 0.84 | 0.231 ± 0.233 | 0.214 ± 0.199 | 0.223 ± 0.207 | −0.019 ± 0.202 | 0.335 ± 0.055 | 24 |
| Pastore d’Oropa | 85.43 | 14.99 | 0.78 | 0.339 ± 0.244 | 0.295 ± 0.179 | 0.305 ± 0.186 | −0.063 ± 0.283 | 0.115 ± 0.089 | 45 |
| Pastore della Lessinia e del Lagorai | 90.33 | 10.00 | 0.76 | 0.323 ± 0.200 | 0.319 ± 0.164 | 0.336 ± 0.172 | 0.033 ± 0.273 | 0.108 ± 0.066 | 32 |
| Pastore della Sila | 85.80 | 13.99 | 0.78 | 0.322 ± 0.233 | 0.290 ± 0.178 | 0.301 ± 0.185 | −0.041 ± 0.261 | 0.082 ± 0.073 | 32 |
| Piccolo Levriero Italiano | 87.89 | 29.88 | 0.79 | 0.260 ± 0.179 | 0.284 ± 0.179 | 0.289 ± 0.182 | 0.085 ± 0.216 | 0.266 ± 0.064 | 72 |
| Portuguese Water Dog | 84.85 | 18.92 | 0.78 | 0.269 ± 0.192 | 0.292 ± 0.182 | 0.300 ± 0.187 | 0.087 ± 0.255 | 0.241 ± 0.073 | 44 |
| Segugio Italiano a Pelo Forte | 92.74 | 16.00 | 0.76 | 0.323 ± 0.186 | 0.321 ± 0.162 | 0.331 ± 0.167 | 0.020 ± 0.226 | 0.107 ± 0.040 | 71 |
| Segugio Italiano a Pelo Raso | 91.02 | 16.00 | 0.77 | 0.313 ± 0.187 | 0.313 ± 0.166 | 0.323 ± 0.172 | 0.024 ± 0.222 | 0.131 ± 0.057 | 64 |
| Spinone Italiano | 88.70 | 17.99 | 0.77 | 0.303 ± 0.193 | 0.303 ± 0.171 | 0.312 ± 0.176 | 0.027 ± 0.226 | 0.157 ± 0.044 | 60 |
| Standard Schnauzer | 87.01 | 13.93 | 0.78 | 0.261 ± 0.186 | 0.298 ± 0.175 | 0.309 ± 0.181 | 0.128 ± 0.290 | 0.258 ± 0.057 | 46 |
| Vizsla | 86.37 | 47.96 | 0.79 | 0.277 ± 0.189 | 0.281 ± 0.181 | 0.284 ± 0.183 | 0.023 ± 0.162 | 0.227 ± 0.061 | 66 |
| Volpino Italiano | 88.76 | 15.96 | 0.78 | 0.283 ± 0.191 | 0.293 ± 0.173 | 0.302 ± 0.179 | 0.053 ± 0.241 | 0.206 ± 0.112 | 44 |
| Wirehaired Pointing Griffon | 87.92 | 17.99 | 0.77 | 0.294 ± 0.191 | 0.305 ± 0.174 | 0.314 ± 0.179 | 0.046 ± 0.237 | 0.182 ± 0.051 | 54 |
| * ROH Category (Mb) | Istrian Shorthaired Hound | Segugio Italiano a Pelo Forte | Segugio Italiano a Pelo Raso | Bracco Italiano | Lagotto Romagnolo | |
|---|---|---|---|---|---|---|
| NROH | all | 186.42 | 151.38 | 157.56 | 210.89 | 167.11 |
| 0.5–2.5 | 159.96 | 133.13 | 134.75 | 152.56 | 127.17 | |
| 2.5–5.0 | 16.98 | 8.87 | 9.81 | 26.56 | 17.50 | |
| >5.0 | 10.59 | 10.60 | 13.00 | 31.78 | 22.44 | |
| LROH (Mb) | all | 1.58 | 1.69 | 1.99 | 2.81 | 2.70 |
| 0.5–2.5 | 0.96 | 0.86 | 0.89 | 0.99 | 0.96 | |
| 2.5–5.0 | 3.41 | 3.47 | 3.47 | 3.61 | 3.52 | |
| >5.0 | 8.71 | 11.30 | 12.26 | 10.87 | 11.88 | |
| SROH (Mb) | all | 294.79 | 255.54 | 313.59 | 592.59 | 450.82 |
| 0.5–2.5 | 153.58 | 114.38 | 120.10 | 151.37 | 122.50 | |
| 2.5–5.0 | 57.90 | 30.79 | 34.09 | 95.88 | 61.60 | |
| >5.0 | 92.20 | 119.78 | 159.40 | 345.34 | 266.72 | |
| FROH | all | 0.12 | 0.11 | 0.13 | 0.25 | 0.19 |
| 0.5–2.5 | 0.06 | 0.05 | 0.05 | 0.06 | 0.05 | |
| 2.5–5.0 | 0.02 | 0.01 | 0.01 | 0.04 | 0.03 | |
| >5.0 | 0.04 | 0.05 | 0.07 | 0.14 | 0.11 |
| Chr. | Start | End | Protein-Coding Genes | Phenotype | Reference |
|---|---|---|---|---|---|
| 10 | 7658201 | 8019503 | WIF1, LEMD3, MSRB3 | Ear morphology in pigs and dogs (WIF1) | [32,33] |
| 14 | 3249578 | 3503246 | LRGUK, EXOC4 | Diabetes and fasting glucose in human | [34] |
| 22 | 2264410 | 2975487 | SETDB2, CAB39L, CDADC1, MLNR, FNDC3A | Pointing vs. herding dog breeds (MLNR) | [35] |
| 28 | 24727970 | 25175110 | NHLRC2, ADRB1, CCDC186, TDRD1, VWA2, AFAP1L2 | Under strong selection in sport-hunting breeds dog (ADRB1) | [36] |
| 30 | 1363393 | 1523168 | RYR3 | Under strong selection in sport-hunting dog breeds | [36] |
| SNP | Chr. | Position | |iHS| | logP | Gene |
|---|---|---|---|---|---|
| BICF2G630421358 | 15 | 57356458 | 4.854 | 5.917 | No annotated genes in this region. |
| BICF2G630525206 | 14 | 29064450 | 4.564 | 5.299 | Intron 20 of the DGKB gene. |
| BICF2G630167445 | 4 | 36002909 | 4.374 | 4.915 | Upstream gene variant of the gene RGS14 (3,6 kbp), 5 prime UTR variant of LMAN2, downstream variant of the gene F12 (4.9 kbp). |
| BICF2S23659818 | 31 | 31128590 | 4.263 | 4.695 | A transcript variant of non-coding RNA gene ENSCAFG00000043029, downstream of the genes cfa-mir-802 (123.3 kbp) and SETD4 (22.0 kbp). |
| BICF2P842400 | 1 | 53824762 | 4.226 | 4.624 | 22.8 kbp upstream of the gene PDE10A. |
| BICF2P722838 | 1 | 53834552 | 4.226 | 4.624 | 32.6 kbp upstream of the gene PDE10A. |
| Breed | Average F (%) | ΔF (%) | Reference |
|---|---|---|---|
| Istrian shorthaired hound | 0.42 | 0.45 | this study |
| Italian pop. of Basset hound | 1.10 | - | [37] |
| Alpine Dachsbrake | 2.26 | - | [39] |
| Finnishound | 3.90 | - | [40] |
| Bracco Italiano | 4.10 | - | [37] |
| Bavarian Mountainhound | 4.5 | 0.69 | [41] |
| Finnishound | 4.60 | - | [42] |
| Braque Francais type Pyrenees | 5.19 | - | [43] |
| German Dachshund | 5.23 | - | [44] |
| Hannoveraner Jagdhund | 6.80 | 0.98 | [41] |
| Tiroler Jagdhund | 9.50 | 1.88 | [41] |
| Polish hunting dog | 11.51 | - | [45] |
| Norwegian Lundehund | 33.00 | 4.00 | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djurkin Kušec, I.; Bošković, I.; Zorc, M.; Gvozdanović, K.; Škorput, D.; Dovč, P.; Kušec, G. Genomic Characterization of the Istrian Shorthaired Hound. Animals 2020, 10, 2013. https://doi.org/10.3390/ani10112013
Djurkin Kušec I, Bošković I, Zorc M, Gvozdanović K, Škorput D, Dovč P, Kušec G. Genomic Characterization of the Istrian Shorthaired Hound. Animals. 2020; 10(11):2013. https://doi.org/10.3390/ani10112013
Chicago/Turabian StyleDjurkin Kušec, Ivona, Ivica Bošković, Minja Zorc, Kristina Gvozdanović, Dubravko Škorput, Peter Dovč, and Goran Kušec. 2020. "Genomic Characterization of the Istrian Shorthaired Hound" Animals 10, no. 11: 2013. https://doi.org/10.3390/ani10112013
APA StyleDjurkin Kušec, I., Bošković, I., Zorc, M., Gvozdanović, K., Škorput, D., Dovč, P., & Kušec, G. (2020). Genomic Characterization of the Istrian Shorthaired Hound. Animals, 10(11), 2013. https://doi.org/10.3390/ani10112013









