Quantification of Myxococcus xanthus Aggregation and Rippling Behaviors: Deep-Learning Transformation of Phase-Contrast into Fluorescence Microscopy Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Time-Lapse Imaging
2.3. Image Processing
2.4. Cell Tracking
2.5. Image Segmentation
2.6. Network Architectures and Learning Algorithm
2.7. Evaluation Metrics
2.7.1. Mean Square Error (MSE) and Structural Similarity Index Measure (SSIM)
2.7.2. Aggregate Comparison
2.7.3. Rippling Wavelength Detection
3. Results
3.1. Fluorescent Images Outperform Phase-Contrast Images in Aggregate Segmentation
3.2. Histogram Equalization (HE) Helps Visualize Streams on Fluorescent Images
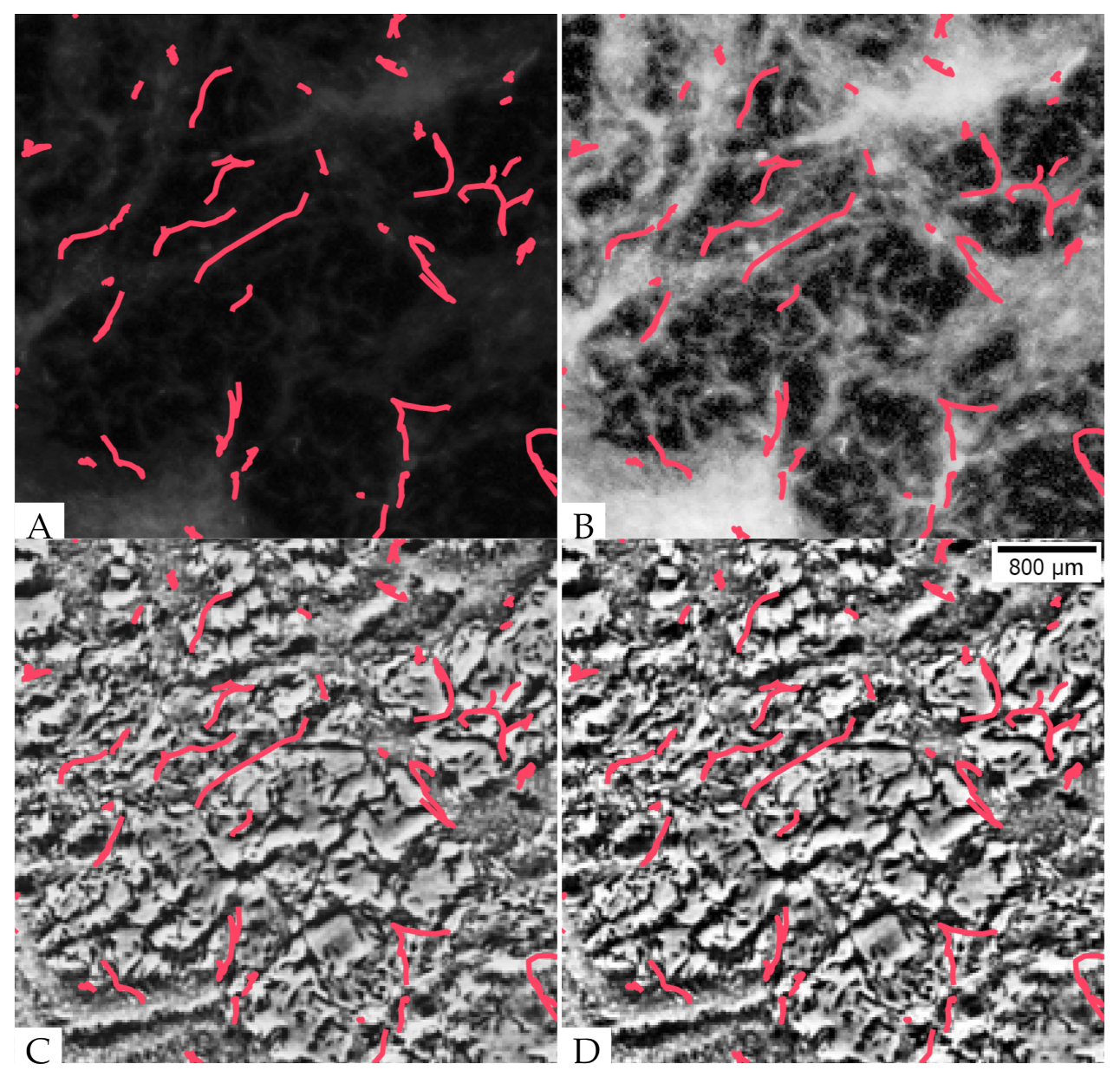
3.3. Deep Neural Network Synthesizes Fluorescent Images with Aggregates and Streams
3.4. The Synthesized Images Show Good Global Agreement with the Real Fluorescent Images
3.5. The Aggregates Segmented from the Synthesized Images Show Good Agreement in Their Positions and Sizes but Some Variability in Aggregate Boundaries
3.6. With Further Training, the Model Can Be Used to Convert Phase-Contrast Images of Rippling and Estimate Its Wavelength
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MSE | Mean Square Error |
| SSIM | Structural Similarity Index Measure |
| HE | Histogram Equalization |
| CLAHE | Contrast Limited Adaptive Histogram Equalization |
| DFT | Discrete Fourier Transformation |
| GAN | Generative Adversarial Network |
| GFP | Green Fluorescent Protein |
References
- Hartzell, T. Myxobacteria. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Bretschneider, T.; Othmer, H.G.; Weijer, C.J. Progress and perspectives in signal transduction, actin dynamics, and movement at the cell and tissue level: Lessons from Dictyostelium. Interface Focus 2016, 6, 20160047. [Google Scholar] [CrossRef] [Green Version]
- Aman, A.; Piotrowski, T. Cell migration during morphogenesis. Dev. Biol. 2010, 341, 20–33. [Google Scholar] [CrossRef] [Green Version]
- Solnica-Krezel, L.; Sepich, D.S. Gastrulation: Making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 2012, 28, 687–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theveneau, E.; Mayor, R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 2012, 366, 34–54. [Google Scholar] [CrossRef] [Green Version]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Bretl, D.J.; Kirby, J.R. Molecular mechanisms of signaling in Myxococcus xanthus development. J. Mol. Biol. 2016, 428, 3805–3830. [Google Scholar] [CrossRef]
- Zhang, Y.; Ducret, A.; Shaevitz, J.; Mignot, T. From individual cell motility to collective behaviors: Insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol. Rev. 2012, 36, 149–164. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Jiang, Y.; Kaiser, A.D.; Alber, M. Self-organization in bacterial swarming: Lessons from myxobacteria. Phys. Biol. 2011, 8, 055003. [Google Scholar] [CrossRef] [PubMed]
- Shimkets, L.J.; Kaiser, D. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 1982, 152, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.; Kaiser, D. Cell behavior in traveling wave patterns of myxobacteria. Proc. Natl. Acad. Sci. USA 2001, 98, 14907–14912. [Google Scholar] [CrossRef] [Green Version]
- Igoshin, O.A.; Mogilner, A.; Welch, R.D.; Kaiser, D.; Oster, G. Pattern formation and traveling waves in myxobacteria: Theory and modeling. Proc. Natl. Acad. Sci. USA 2001, 98, 14913–14918. [Google Scholar] [CrossRef] [Green Version]
- Berleman, J.E.; Chumley, T.; Cheung, P.; Kirby, J.R. Rippling is a predatory behavior in Myxococcus xanthus. J. Bacteriol. 2006, 188, 5888–5895. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Vaksman, Z.; Litwin, D.B.; Shi, P.; Kaplan, H.B.; Igoshin, O.A. The Mechanistic Basis of Myxococcus xanthus Rippling Behavior and Its Physiological Role during Predation. PLoS Comput. Biol. 2012, 8, e1002715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotter, C.R.; Schüttler, H.B.; Igoshin, O.A.; Shimkets, L.J. Data-driven modeling reveals cell behaviors controlling self-organization during Myxococcus xanthus development. Proc. Natl. Acad. Sci. USA 2017, 114, E4592–E4601. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Cotter, C.R.; Lyu, Z.; Shimkets, L.J.; Igoshin, O.A. Data-driven models reveal mutant cell behaviors important for myxobacterial aggregation. Msystems 2020, 5, e00518-20. [Google Scholar] [CrossRef]
- Otaki, T. Artifact halo reduction in phase contrast microscopy using apodization. Opt. Rev. 2000, 7, 119–122. [Google Scholar] [CrossRef]
- Maurer, C.; Jesacher, A.; Bernet, S.; Ritsch-Marte, M. Phase contrast microscopy with full numerical aperture illumination. Opt. Express 2008, 16, 19821–19829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Icha, J.; Weber, M.; Waters, J.C.; Norden, C. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays 2017, 39, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.Y.; Huang, X.; Yu, J.; Wang, T.C.; Mallya, A. Generative adversarial networks for image and video synthesis: Algorithms and applications. Proc. IEEE 2021, 109, 839–862. [Google Scholar]
- Pang, Y.; Lin, J.; Qin, T.; Chen, Z. Image-to-Image Translation: Methods and Applications. arXiv 2021, arXiv:2101.08629. [Google Scholar]
- Suárez, P.L.; Sappa, A.D.; Vintimilla, B.X. Infrared Image Colorization Based on a Triplet DCGAN Architecture. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 212–217. [Google Scholar] [CrossRef]
- Zhang, H.; Sindagi, V.; Patel, V.M. Image de-raining using a conditional generative adversarial network. IEEE Trans. Circuits Syst. Video Technol. 2019, 30, 3943–3956. [Google Scholar] [CrossRef] [Green Version]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the Proceedings—30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2242–2251. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Chen, Q.; Koltun, V. Photographic image synthesis with cascaded refinement networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 1511–1520. [Google Scholar]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-Resolution Image Synthesis and Semantic Manipulation with Conditional GANs. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar] [CrossRef] [Green Version]
- Park, T.; Liu, M.Y.; Wang, T.C.; Zhu, J.Y. Semantic image synthesis with spatially-adaptive normalization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2337–2346. [Google Scholar]
- Curtis, P.D.; Taylor, R.G.; Welch, R.D.; Shimkets, L.J. Spatial Organization of Myxococcus xanthus during Fruiting Body Formation. J. Bacteriol. 2007, 189, 9126–9130. [Google Scholar] [CrossRef] [Green Version]
- Otsu, N.; Smith, P.L.; Reid, D.B.; Environment, C.; Palo, L.; Alto, P.; Smith, P.L. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, C, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Vondrick, C.; Fowlkes, C.C.; Ramanan, D. Do we need more training data? Int. J. Comput. Vis. 2016, 119, 76–92. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.; Boynton, T.O.; Pham, D.; Shimkets, L.J. Fatty Acids from Membrane Lipids Become Incorporated into Lipid Bodies during Myxococcus xanthus Differentiation. PLoS ONE 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 270–279. [Google Scholar]
- Kaiser, D.; Welch, R. Dynamics of fruiting body morphogenesis. J. Bacteriol. 2004, 186, 919–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolgemuth, C.; Hoiczyk, E.; Kaiser, D.; Oster, G. How Myxobacteria Glide. Curr. Biol. 2002, 12, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.; Kaiser, D. Gliding motility and polarized slime secretion. Mol. Microbiol. 2007, 63, 454–467. [Google Scholar] [CrossRef]






| MSE | SSIM | ||||||||
| Model | Density (cells/mL) | Type | Value | SD | Difference | Value | SD | Difference | |
| pix2pixHD | training | 0.019 ± 0.014 | 6.8 | 6.8% | −0.001 ± 0.009 | 0.736 ± 0.111 | 2.2 | 0.008 ± 0.032 | |
| pix2pixHD-HE | training | 0.018 ± 0.015 | 6.2 | 6.7% | 0.744 ± 0.117 | 2.1 | |||
| pix2pixHD | test | 0.026 ± 0.015 | 6.9 | 8.1% | −0.002 ± 0.008 | 0.680 ± 0.102 | 2.2 | 0.008 ± 0.024 | |
| pix2pixHD-HE | test | 0.025 ± 0.012 | 7.0 | 7.8% | 0.687 ± 0.103 | 2.1 | |||
| pix2pixHD | training | 0.017 ± 0.013 | 4.1 | 6.6% | 0.000 ± 0.006 | 0.721 ± 0.128 | 1.9 | −0.001 ± 0.032 | |
| pix2pixHD-HE | training | 0.017 ± 0.011 | 4.3 | 6.6% | 0.720 ± 0.133 | 2.0 | |||
| pix2pixHD | test | 0.031 ± 0.022 | 5.3 | 8.7% | −0.001 ± 0.008 | 0.662 ± 0.111 | 2.2 | 0.002 ± 0.025 | |
| pix2pixHD-HE | test | 0.030 ± 0.023 | 4.8 | 8.6% | 0.664 ± 0.103 | 2.1 | |||
| pix2pixHD | test | 0.119 ± 0.035 | 22.6 | 17.3% | -0.032 ± 0.007 | 0.420 ± 0.069 | 4.5 | 0.044 ± 0.010 | |
| pix2pixHD-HE | test | 0.087 ± 0.029 | 16.6 | 14.8% | 0.465 ± 0.074 | 4.0 | |||
| MSE | SSIM | ||||||||
| Model | Density (cells/mL) | Type | Value | SD | Difference | Value | SD | Difference | |
| pix2pixHD | training | 0.101 ± 0.042 | 3.3 | 15.9% | −0.005 ± 0.010 | 0.458 ± 0.115 | 2.2 | 0.019 ± 0.025 | |
| pix2pixHD-HE | training | 0.096 ± 0.043 | 3.2 | 15.5% | 0.478 ± 0.123 | 2.2 | |||
| pix2pixHD | test | 0.161 ± 0.074 | 3.7 | 20.1% | −0.006 ± 0.009 | 0.385 ± 0.125 | 2.4 | 0.016 ± 0.017 | |
| pix2pixHD-HE | test | 0.155 ± 0.079 | 3.6 | 19.7% | 0.401 ± 0.131 | 2.3 | |||
| pix2pixHD | training | 0.098 ± 0.076 | 2.9 | 15.7% | −0.003 ± 0.006 | 0.487 ± 0.134 | 2.2 | 0.012 ± 0.015 | |
| pix2pixHD-HE | training | 0.095 ± 0.078 | 2.7 | 15.4% | 0.499 ± 0.139 | 2.1 | |||
| pix2pixHD | test | 0.182 ± 0.064 | 2.5 | 21.3% | 0.003 ± 0.010 | 0.338 ± 0.107 | 2.3 | −0.001 ± 0.019 | |
| pix2pixHD-HE | test | 0.184 ± 0.066 | 2.5 | 21.5% | 0.336 ± 0.114 | 2.3 | |||
| pix2pixHD | test | 0.123 ± 0.051 | 5.2 | 17.6% | −0.001 ± 0.005 | 0.472 ± 0.098 | 3.0 | 0.005 ± 0.008 | |
| pix2pixHD-HE | test | 0.122 ± 0.049 | 5.1 | 17.5% | 0.477 ± 0.094 | 2.9 | |||
| Cell Density (cells/mL) | Image Type | Displacement (m) | Relative Area | Precision | Recall |
|---|---|---|---|---|---|
| synthesized | 4.2 ± 3.1 | 1.04 ± 0.31 | 0.82 | 0.88 | |
| phase-contrast | 10.0 ± 7.3 | 1.18 ± 0.40 | 0.69 | 0.81 | |
| synthesized | 4.4 ± 3.3 | 1.18 ± 0.31 | 0.82 | 0.95 | |
| phase-contrast | 9.9 ± 10.6 | 1.27 ± 0.42 | 0.70 | 0.88 | |
| synthesized | 7.1 ± 5.1 | 0.90 ± 0.24 | 0.90 | 0.83 | |
| phase-contrast | 32.6 ± 16.6 | 0.68 ± 0.43 | 0.48 | 0.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Comstock, J.A.; Cotter, C.R.; Murphy, P.A.; Nie, W.; Welch, R.D.; Patel, A.B.; Igoshin, O.A. Quantification of Myxococcus xanthus Aggregation and Rippling Behaviors: Deep-Learning Transformation of Phase-Contrast into Fluorescence Microscopy Images. Microorganisms 2021, 9, 1954. https://doi.org/10.3390/microorganisms9091954
Zhang J, Comstock JA, Cotter CR, Murphy PA, Nie W, Welch RD, Patel AB, Igoshin OA. Quantification of Myxococcus xanthus Aggregation and Rippling Behaviors: Deep-Learning Transformation of Phase-Contrast into Fluorescence Microscopy Images. Microorganisms. 2021; 9(9):1954. https://doi.org/10.3390/microorganisms9091954
Chicago/Turabian StyleZhang, Jiangguo, Jessica A. Comstock, Christopher R. Cotter, Patrick A. Murphy, Weili Nie, Roy D. Welch, Ankit B. Patel, and Oleg A. Igoshin. 2021. "Quantification of Myxococcus xanthus Aggregation and Rippling Behaviors: Deep-Learning Transformation of Phase-Contrast into Fluorescence Microscopy Images" Microorganisms 9, no. 9: 1954. https://doi.org/10.3390/microorganisms9091954
APA StyleZhang, J., Comstock, J. A., Cotter, C. R., Murphy, P. A., Nie, W., Welch, R. D., Patel, A. B., & Igoshin, O. A. (2021). Quantification of Myxococcus xanthus Aggregation and Rippling Behaviors: Deep-Learning Transformation of Phase-Contrast into Fluorescence Microscopy Images. Microorganisms, 9(9), 1954. https://doi.org/10.3390/microorganisms9091954








