Historical Differentiation and Recent Hybridization in Natural Populations of the Nematode-Trapping Fungus Arthrobotrys oligospora in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Identification of Tandem Repeat Loci and Development of Amplification Primers
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Data Analysis
2.5. Intraspecific Phenotypic Variation
2.5.1. Comparison of Mycelial Growth and Conidial Shape
2.5.2. Trap Formation and Bioassay
2.5.3. Nematicidal Activity of Fermentation Broth
2.5.4. Statistical Analysis
3. Results
3.1. Genetic Diversity of the Chinese Samples of A. oligospora Detected by STRs and MLST
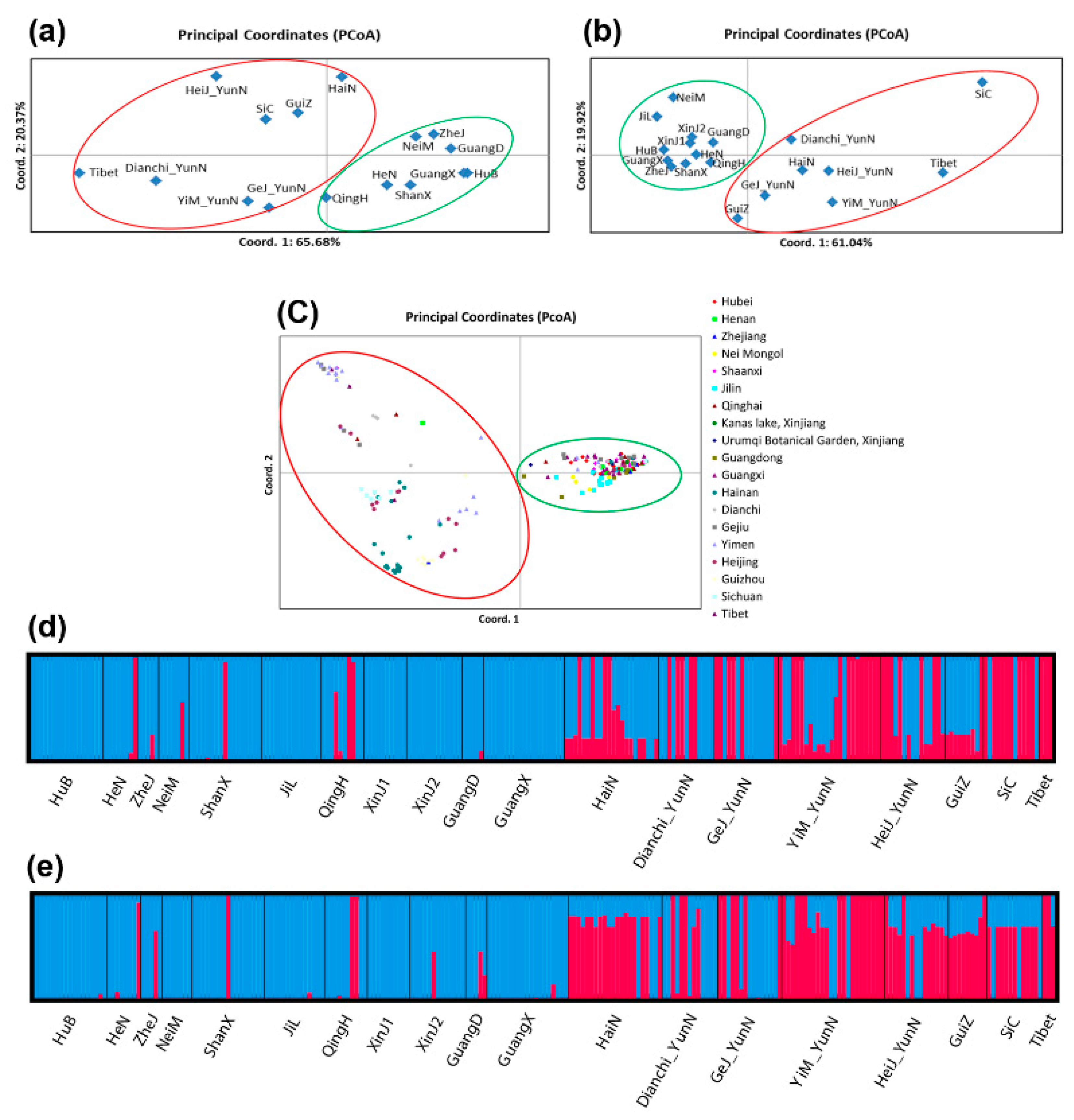
3.2. Genetic Differentiation and Population Structure
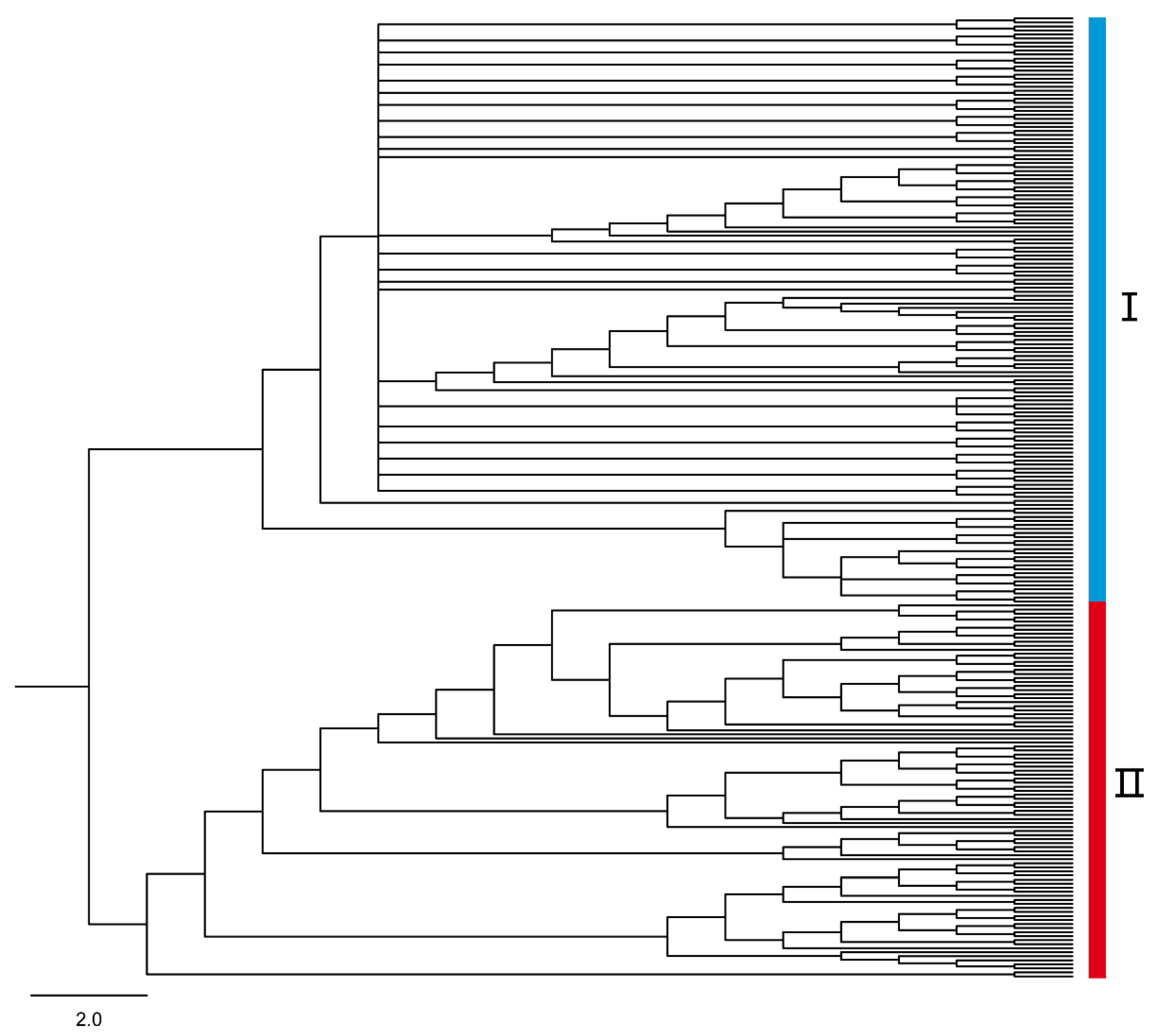
3.3. Phylogenetic Divergence
3.4. Clonality and Recombination
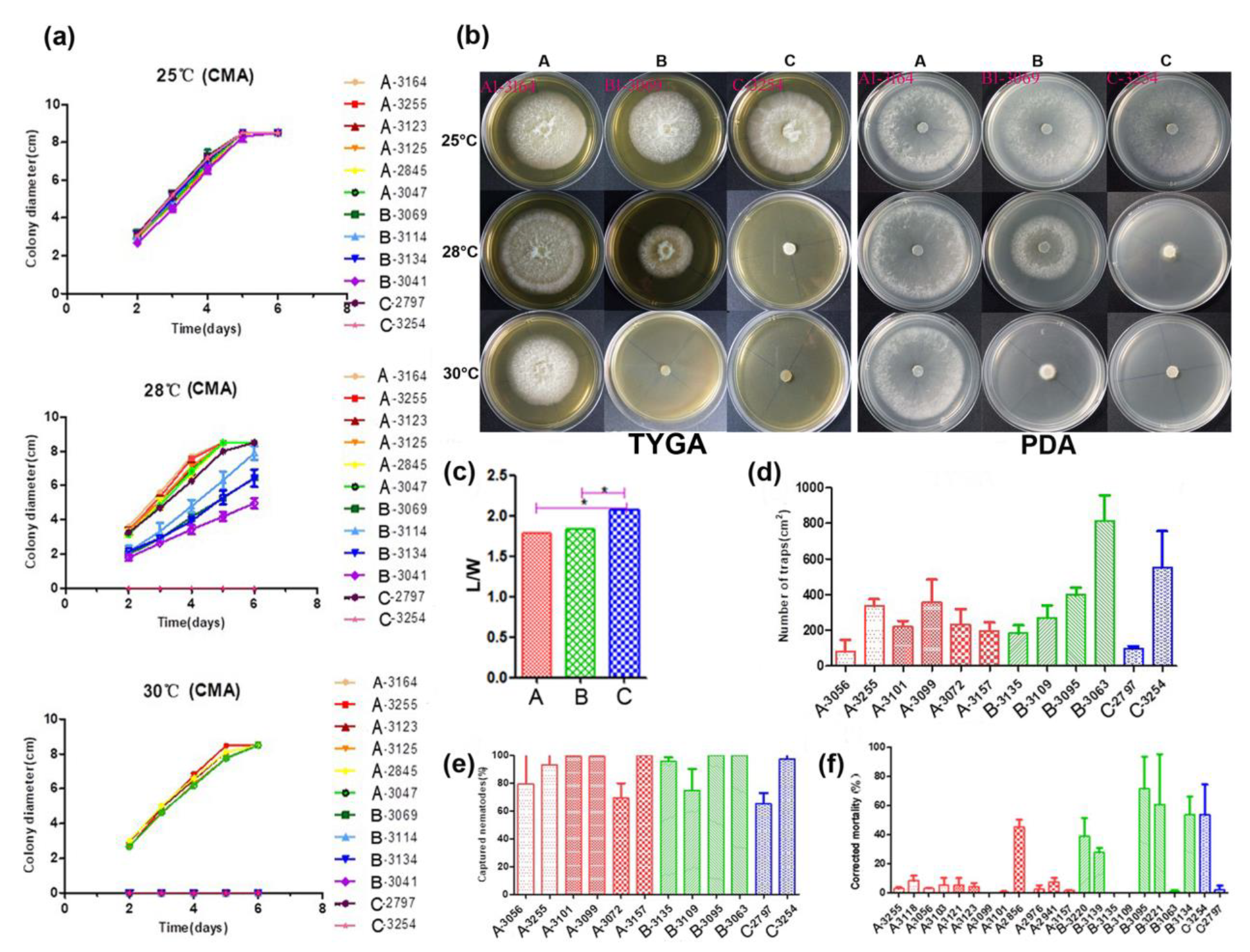
3.5. Phenotypical Characterization
3.5.1. Conidial Morphology
3.5.2. Mycelial Growth
3.5.3. Conidial Yield, Trap Formation and Nematode-Trapping Bioassay
4. Discussion
4.1. Development of Novel STRs for NTF
4.2. Historical Population Differentiation
4.3. Recent Hybridization and Recombination
4.4. Intraspecies Diversification
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abad, P.; Gouzy, J.; Aury, J.-M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotech. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Giannakou, I.O.; Anastasiadis, I.A.; Gowen, S.R.; Prophetou-Athanasiadou, D. Effects of a non-chemical nematicide combined with soil solarization for the control of root-knot nematodes. Crop Prot. 2007, 26, 1644–1654. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Li, H.; Wang, R.; Zhang, K.Q.; Xu, J. Fungi-Nematode Interactions: Diversity, Ecology, and Biocontrol Prospects in Agriculture. J. Fungi 2020, 6, 206. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xiang, M.; Liu, X. Nematode-Trapping Fungi. Microbiol. Spectr. 2017, 5, FUNK-0022-2016. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.-G.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and Proteomic Analyses of the Fungus Arthrobotrys oligospora Provide Insights into Nematode-Trap Formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, K.-Q.; Hyde, K. The Ecology of Nematophagous Fungi in Natural Environments. In Nematode-Trapping Fungi; Zhang, K.-Q., Hyde, K.D., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 211–229. [Google Scholar] [CrossRef]
- Li, J.; Zou, C.; Xu, J.; Ji, X.; Niu, X.; Yang, J.; Huang, X.; Zhang, K.Q. Molecular mechanisms of nematode-nematophagous microbe interactions: Basis for biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 2015, 53, 67–95. [Google Scholar] [CrossRef]
- Gray, N.F. Ecology of nematophagous fungi: Distribution and habitat. Ann. Appl. Biol. 1983, 102, 501–509. [Google Scholar] [CrossRef]
- Niu, X.-M.; Zhang, K.-Q. Arthrobotrys oligospora: A model organism for understanding the interaction between fungi and nematodes. Mycology 2011, 2, 59–78. [Google Scholar] [CrossRef]
- Waghorn, T.S.; Leathwick, D.M.; Chen, L.Y.; Gray, R.A.; Skipp, R.A. Influence of nematophagous fungi, earthworms and dung burial on development of the free-living stages of Ostertagia (Teladorsagia) circumcincta in New Zealand. Vet. Parasitol. 2002, 104, 119–129. [Google Scholar] [CrossRef]
- Gray, N. Nematophagous fungi with particular reference to their ecology. Biol. Rev. Cambri. Philosophi. Soc. 1987, 62, 245–304. [Google Scholar] [CrossRef]
- Pathak, E.; Campos–Herrera, R.; El–Borai, F.E.; Duncan, L.W. Spatial relationships between entomopathogenic nematodes and nematophagous fungi in Florida citrus orchards. J. Inverteb. Pathol. 2017, 144, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Jaffee, B.A.; Strong, D.R.; Muldoon, A.E. Nematode-trapping fungi of a natural shrubland: Tests for food chain involvement. Mycologia 2018, 88, 554–564. [Google Scholar] [CrossRef]
- Deng, W.; Wang, J.L.; Scott, M.B.; Fang, Y.H.; Liu, S.R.; Yang, X.Y.; Xiao, W. Sampling methods affect Nematode-Trapping Fungi biodiversity patterns across an elevational gradient. BMC Microbiol. 2020, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Strom, N.; Hu, W.; Haarith, D.; Chen, S.; Bushley, K. Interactions between soil properties, fungal communities, the soybean cyst nematode, and crop yield under continuous corn and soybean monoculture. Appl. Soil Ecol. 2020, 147, 103388. [Google Scholar] [CrossRef]
- Li, Y.; Hyde, K.D.; Jeewon, R.; Cai, L.; Vijaykrishna, D.; Zhang, K. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia 2005, 97, 1034–1046. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rubner, A. Revision of predacious hyphomycetes in the Dactylella-Monacrosporium complex. Stud. Mycol. 1996, 39, 1–134. [Google Scholar]
- Scholler, M.; Hagedorn, G.; Rubner, A. A reevaluation of predatory orbiliaceous fungi. II. A new generic concept. Sydowia 1999, 51, 89–113. [Google Scholar]
- Yang, E.; Xu, L.; Yang, Y.; Zhang, X.; Xiang, M.; Wang, C.; An, Z.; Liu, X. Origin and evolution of carnivorism in the Ascomycota (fungi). Proc. Natl. Acad. Sci. USA 2012, 109, 10960–10965. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, E.C.; An, Z.Q.; Liu, X.Z. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. USA 2007, 104, 8379–8384. [Google Scholar] [CrossRef]
- Yang, C.T.; Vidal-Diez de Ulzurrun, G.; Goncalves, A.P.; Lin, H.C.; Chang, C.W.; Huang, T.Y.; Chen, S.A.; Lai, C.K.; Tsai, I.J.; Schroeder, F.C.; et al. Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 6762–6770. [Google Scholar] [CrossRef]
- Liu, M.; Cheng, X.; Wang, J.; Tian, D.; Tang, K.; Xu, T.; Zhang, M.; Wang, Y.; Wang, M. Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora. Int. J. Biol. Macromol. 2020, 164. [Google Scholar] [CrossRef]
- Kuo, T.H.; Yang, C.T.; Chang, H.Y.; Hsueh, Y.P.; Hsu, C.C. Nematode-Trapping Fungi Produce Diverse Metabolites during Predator-Prey Interaction. Metabolites 2020, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Yu, Z.; Yang, J.; Xu, J.; Zhang, Y.; Liu, S.; Zou, C.; Li, J.; Liang, L.; Zhang, K.Q. Expansion of Adhesion Genes Drives Pathogenic Adaptation of Nematode-Trapping Fungi. iScience 2020, 23, 101057. [Google Scholar] [CrossRef]
- Basso, M.F.; Lourenco-Tessutti, I.T.; Mendes, R.A.G.; Pinto, C.E.M.; Bournaud, C.; Gillet, F.X.; Togawa, R.C.; de Macedo, L.L.P.; de Almeida Engler, J.; Grossi-de-Sa, M.F. MiDaf16-like and MiSkn1-like gene families are reliable targets to develop biotechnological tools for the control and management of Meloidogyne incognita. Sci. Rep. 2020, 10, 6991. [Google Scholar] [CrossRef]
- Liang, L.M.; Zou, C.G.; Xu, J.; Zhang, K.Q. Signal pathways involved in microbe-nematode interactions provide new insights into the biocontrol of plant-parasitic nematodes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180317. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Dong, L.; He, R.; Wang, Q.; Chen, Y.; Liangjian, Q.; Zhang, Y.-A. Comparative genomic analyses reveal the features for adaptation to nematodes in fungi. DNA Res. 2018, 25, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.P.; Gronquist, M.R.; Schwarz, E.M.; Nath, R.D.; Lee, C.H.; Gharib, S.; Schroeder, F.C.; Sternberg, P.W. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. eLife 2017, 6, e20023. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, G.H.; Zou, C.G.; Ji, X.L.; Liu, T.; Zhao, P.J.; Liang, L.M.; Xu, J.P.; An, Z.Q.; Zheng, X.; et al. Bacteria can mobilize nematode-trapping fungi to kill nematodes. Nat. Commun. 2014, 5, 5776. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, Z.-F.; Xu, J.; Zhang, K.-Q. Divergence and dispersal of the nematode-trapping fungus Arthrobotrys oligospora from China. Environ. Microbiol. Rep. 2011, 3, 763–773. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, M.; Xu, J.; Cao, Y.; Zhang, K.Q.; Yu, Z.F. Genetic diversity and recombination in natural populations of the nematode-trapping fungus Arthrobotrys oligospora from China. Ecol. Evol. 2013, 3, 312–325. [Google Scholar] [CrossRef]
- Borstnik, B.; Pumpernik, D. Tandem repeats in protein coding regions of primate genes. Genome Res. 2002, 12, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Dallas, J.F. Estimation of microsatellite mutation rates in recombinant inbred strains of mouse. Mammalian Genome 1992, 3, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Karaoglu, H.; Lee, C.M.; Meyer, W. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 2005, 22, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhou, Y.; Price, M.; Song, Z. Genome-wide characterization of microsatellite DNA in fishes: Survey and analysis of their abundance and frequency in genome-specific regions. BMC Genomics 2021, 22, 421. [Google Scholar] [CrossRef] [PubMed]
- Sasseron, G.R.; Benchimol-Reis, L.L.; Perseguini, J.; Paulino, J.F.C.; Bajay, M.M.; Carbonell, S.A.M.; Chiorato, A.F. Fusarium oxysporum f. sp. phaseoli genetic variability assessed by new developed microsatellites. Genet. Mol. Biol. 2020, 43, e20190267. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yu, J.; Gu, C.; Nie, Y.; Chen, Z.; Yin, X.; Liu, Y. De novo sequencing and transcriptome analysis of Ustilaginoidea virens by using Illumina paired-end sequencing and development of simple sequence repeat markers. Gene 2014, 547, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, G.; Fang, M.; Deng, C.; Xu, J. Comparative Analyses of Mitochondrial Genomes Provide Evolutionary Insights Into Nematode-Trapping Fungi. Front. Microbiol. 2020, 11, 617. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.A., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Deigo, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mo, M.; Yang, L.; Mi, F.; Cao, Y.; Liu, C.; Tang, X.; Wang, P.; Xu, J. Exploring the Species Diversity of Edible Mushrooms in Yunnan, Southwestern China, by DNA Barcoding. J. Fungi 2021, 7, 310. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Pfister, D.; Liftik, M. Two Arthrobotrys anamorphs from Orbilia auricolor. Mycologia 1995, 87, 684–688. [Google Scholar] [CrossRef]
- Mo, M.H.; Huang, X.W.; Zhou, W. Arthrobotrys yunnanensis sp. nov., the fourth anamorph of Orbilia auricolor. Fungal Divers. 2005, 18, 107–115. [Google Scholar]
- Shenoy, B.D.; Jeewon, R.; Hyde, K.; Hyde, R. Impact of DNA sequence-data on the taxonomy of anamorphic fungi. Fungal Divers. 2007, 26, 1–54. [Google Scholar]
- Agapow, P.M.; Burt, A. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 2001, 1, 101–102. [Google Scholar] [CrossRef]
- Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhou, D.; Zhao, Y.; Li, D.; Niu, X.; Zhang, K.Q.; Yang, J. Protein Kinase Ime2 Is Required for Mycelial Growth, Conidiation, Osmoregulation, and Pathogenicity in Nematode-Trapping Fungus Arthrobotrys oligospora. Front. Microbiol. 2019, 10, 3065. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Z.; Zhang, G.; Yang, L.; Ma, N.; Li, Q.; Ma, Y.; Niu, X.; Zhang, K.Q.; Yang, J. Characterization and functional analysis of calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2019, 103, 819–832. [Google Scholar] [CrossRef]
- Barrière, A.; Félix, M.-A. Isolation of C. elegans and related nematodes. In WormBook; 2014; pp. 1–19. Available online: https://europepmc.org/article/med/24803426 (accessed on 15 July 2020).
- Botstein, D.; White, R.L.; Skolnick, M.H.; Davis, R. Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Smith, M.E.; Jaffee, B.A. PCR primers with enhanced specificity for nematode-trapping fungi (Orbiliales). Microb. Ecol. 2009, 58, 117–128. [Google Scholar] [CrossRef]
- Bailey, F.; Gray, N.F. The comparison of isolation techniques for nematophagous fungi from soil. Ann. Appl. Biol. 1989, 114, 125–132. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Feng, B.; Yang, Z. Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Divers. 2018, 40, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Wang, R.; Li, X.; Peng, B.; Yang, G.; Zhang, K.-Q.; Zhang, Y.; Xu, J. Genetic Diversity and Azole Resistance Among Natural Aspergillus fumigatus Populations in Yunnan, China. Microb. Ecol. 2021, 10. [Google Scholar] [CrossRef]
- Zhou, D.; Korfanty, G.A.; Mo, M.; Wang, R.; Li, X.; Li, H.; Li, S.; Wu, J.Y.; Zhang, K.Q.; Zhang, Y.; et al. Extensive Genetic Diversity and Widespread Azole Resistance in Greenhouse Populations of Aspergillus fumigatus in Yunnan, China. mSphere 2021, 6, e00066-21. [Google Scholar] [CrossRef]
- Wu, J.-Y.; Guo, H.; Wang, H.-M.; Yi, G.-H.; Zhou, L.-M.; He, X.-W.; Zhang, Y.; Xu, J. Multilocus sequence analyses reveal extensive diversity and multiple origins of fluconazole resistance in Candida tropicalis from tropical China. Sci. Rep. 2017, 7, 42537. [Google Scholar] [CrossRef]
- Fang, D.Z.; Liu, X.L.; Chen, X.R.; Yan, W.R.; He, Y.L.; Cheng, Y.; Chen, J.; Li, Z.M.; Guo, L.T.; Wang, T.H.; et al. Fusarium Species and Fusarium oxysporum Species Complex Genotypes Associated With Yam Wilt in South-Central China. Front. Microbiol. 2020, 11, 1964. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Y.; Yu, Z.; Mi, F.; Liu, C.; Tang, X.; Long, Y.; He, X.; Wang, P.; Xu, J. Structure, Gene Flow, and Recombination among Geographic Populations of a Russula virescens Ally from Southwestern China. PLoS ONE 2013, 8, e73174. [Google Scholar] [CrossRef]
- Xu, J. Fungal Species Concepts in the Genomics Era. Genome 2020, 63, 459–468. [Google Scholar] [CrossRef]
- Guo, T.; Wang, H.C.; Xue, W.Q.; Zhao, J.; Yang, Z.L. Phylogenetic Analyses of Armillaria Reveal at Least 15 Phylogenetic Lineages in China, Seven of Which Are Associated with Cultivated Gastrodia elata. PLoS ONE 2016, 11, e0154794. [Google Scholar] [CrossRef] [PubMed]
- Peintner, U.; Kuhnert-Finkernagel, R.; Wille, V.; Biasioli, F.; Shiryaev, A.; Perini, C. How to resolve cryptic species of polypores: An example in Fomes. IMA Fungus 2019, 10, 17. [Google Scholar] [CrossRef]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef]
- Brown, E.M.; McTaggart, L.R.; Zhang, S.X.; Low, D.E.; Stevens, D.A.; Richardson, S.E. Phylogenetic Analysis Reveals a Cryptic Species Blastomyces gilchristii, sp. nov. within the Human Pathogenic Fungus Blastomyces dermatitidis. PLoS ONE 2013, 8, e59237. [Google Scholar] [CrossRef] [PubMed]
- Ahrén, D.; Faedo, M.; Rajastiekar, B.; Tunlid, A. Low genetic diversity among isolates of the nematode-trapping fungus Duddingtonia flagrans: Evidence for recent worldwide dispersion from a single common ancestor. Mycol. Res. 2004, 108, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Carta, L.; Rehner, S. Morphological variability and molecular phylogeny of the nematophagous fungus Monacrosporium drechsleri. Mycologia 2005, 97, 405–415. [Google Scholar] [CrossRef]
- Savary, R.; Masclaux, F.G.; Wyss, T.; Droh, G.; Cruz Corella, J.; Machado, A.P.; Morton, J.B.; Sanders, I.R. A population genomics approach shows widespread geographical distribution of cryptic genomic forms of the symbiotic fungus Rhizophagus irregularis. ISME J. 2018, 12, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Bracewell, R.; Vanderpool, D.; Good, J.; Six, D. Cascading speciation among mutualists and antagonists in a tree–beetle–fungi interaction. Proc. Roy. Soc. B Biol. Sci. 2018, 285, 20180694. [Google Scholar] [CrossRef]
- Chaliha, C.; Kaladhar, C.; Doley, R.; Verma, P.; Kumar, A.; Kalita, E. Bipartite molecular approach for species delimitation and resolving cryptic speciation of Exobasidium vexans within the Exobasidium genus. Comput. Biol. Chem. 2021, 92, 107496. [Google Scholar] [CrossRef]
- Hartmann, F.E.; Snirc, A.; Cornille, A.; Godé, C.; Touzet, P.; Van Rossum, F.; Fournier, E.; Le Prieur, S.; Shykoff, J.; Giraud, T. Congruent population genetic structures and divergence histories in anther-smut fungi and their host plants Silene italica and the Silene nutans species complex. Mol. Ecol. 2020, 29, 1154–1172. [Google Scholar] [CrossRef]
- St Leger, R.J.; Screen, S.E.; Shams-Pirzadeh, B. Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 2000, 66, 320–324. [Google Scholar] [CrossRef]
- Bidochka, M.J.; Kamp, A.M.; Lavender, T.M.; Dekoning, J.; De Croos, J.N.A. Habitat association in two genetic groups of the insect-pathogenic fungus Metarhizium anisopliae: Uncovering cryptic species? Appl. Environ. Microbiol. 2001, 67, 1335–1342. [Google Scholar] [CrossRef] [PubMed]



| Population | No. Isolates | No. Alleles in Each Locus (No. Private Alleles) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A3 | A5 | A17 | A25 | A51 | A74 | A80 | A83 | A87 | A101 | A103 | A126 | A149 | A154 | A156 | A160 | A177 | A187 | A191 | A192 | Total | ||
| Hubei (HuB) | 17 | 1 | 1 | 3 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 (1) | 2 | 2 | 3 (1) | 3 | 2 | 3 | 1 | 38 (2) |
| Henan (HeN) | 8 | 2 | 1 | 2 (1) | 1 | 1 | 3 | 2 | 2 | 2 | 3 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 3 | 5 | 2 | 45 (1) |
| Zhejiang (ZheJ) | 5 | 1 | 2 | 3 | 1 | 2 (1) | 3 | 2 | 2 | 1 | 4 | 2 | 2 | 1 | 1 | 3 | 1 | 4 | 2 | 3 | 2 | 42 (1) |
| Nei Mongol (NeiM) | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 23 |
| Shaanxi (ShanX) | 17 | 2 | 1 | 3 | 2 | 2 | 4 | 2 | 4 | 2 | 5 | 5 | 4 (1) | 2 | 3 | 3 | 3 | 3 | 3 | 4 | 2 | 59 (1) |
| Jilin (JiL) | 14 | 1 | 1 | 2 | 1 | 1 | 3 | 1 | 2 | 1 | 3 | 2 | 2 | 1 | 3 (1) | 2 | 1 | 1 | 2 | 3 (1) | 1 | 34 (2) |
| Qinghai (QingH) | 10 | 2 | 2 | 3 | 3 | 2 | 4 | 3 | 2 | 2 | 5 | 5 | 3 | 2 | 3 | 3 | 3 | 5 | 3 | 5 | 3 | 63 |
| Kanas lake, Xinjiang (XinJ1) | 10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 21 |
| Urumqi Botanical Garden, Xinjiang (XinJ2) | 13 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | 2 (1) | 2 | 2 | 1 | 2 | 1 | 1 | 32 (1) |
| Guangdong (GuangD) | 5 | 1 | 1 | 4 | 1 | 2 | 2 | 2 | 2 | 2 | 5 | 2 | 3 | 2 | 1 | 3 (1) | 2 | 2 | 2 | 4 | 2 | 45 (1) |
| Guangxi (GuangX) | 19 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | 3 | 2 (1) | 5 (2) | 3 | 2 | 2 | 2 | 2 | 2 (1) | 3 | 2 | 4 (1) | 2 (1) | 44 (6) |
| Hainan (HaiN) | 22 | 1 | 3 | 4 | 5 | 3 | 3 | 3 | 5 | 4 | 4 | 4 | 4 (1) | 6 (2) | 2 | 3 | 2 | 4 | 3 | 5 (2) | 3 | 71 (5) |
| Dianchi lake, Yunnan (Dianchi_YunN) | 13 | 3 | 3 | 2 | 4 | 2 | 3 | 4 | 4 (2) | 4 (1) | 4 | 5 | 6 | 4 | 2 | 3 | 3 | 4 | 3 | 4 (1) | 4 | 71 (4) |
| Gejiu, Yunnan (GeJ_YunN) | 15 | 3 | 2 | 3 | 3 | 4 | 3 | 3 | 3 | 3 | 5 | 5 (1) | 2 | 3 | 3 | 4 (1) | 3 (1) | 4 | 3 (1) | 5 | 3 (1) | 67 (5) |
| Yimen, Yunnan (YiM_YunN) | 24 | 3 | 2 | 4 | 3 | 3 | 4 | 6 (2) | 5 (1) | 3 | 5 (1) | 3 | 4 | 4 | 3 | 4 | 4 (2) | 6 (2) | 3 | 7 (1) | 3 | 79 (9) |
| Heijing, Yunnan (HeiJ_YunN) | 15 | 2 | 5 | 2 | 3 | 3 | 5 | 3 | 5 | 4 | 8 (1) | 4 | 5 | 2 | 4 (1) | 4 | 5 | 5 | 6 | 8 (1) | 5 | 88 (3) |
| Guizhou (GuiZ) | 9 | 2 | 2 | 3 | 1 | 3 (1) | 2 | 2 | 3 | 2 | 3 (1) | 3 | 3 | 2 | 2 | 3 | 2 | 2 | 3 | 2 | 2 | 47 (2) |
| Sichuan (SiC) | 13 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 3 | 2 | 3 | 3 (1) | 2 | 3 | 1 | 2 | 2 | 3 | 2 | 2 | 2 | 44 (1) |
| Tibet | 3 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 (1) | 1 | 2 | 2 | 2 (1) | 2 | 2 | 3 | 1 | 3 (2) | 1 | 2 | 41 (4) |
| Total | 239 | 3 | 5 | 7 | 5 | 9 | 10 | 7 | 10 | 8 | 16 | 13 | 10 | 11 | 6 | 8 | 11 | 12 | 9 | 21 | 7 | 188 (48) |
| Locus | Forward Primer Sequence | Reverse Primer Sequence | Repeat Unit | PIC | Allele Frequency | Availability | Gene Diversity |
|---|---|---|---|---|---|---|---|
| A3 | FAM-GGAGTGGAAGTTAGATTGGAG | GGGGAACTAATTTACTTGCAT | GTG | 0.244 | 0.849 | 0.941 | 0.265 |
| A5 | HEX-CGATGAAGACGTGAGTTAGTT | TGTCTGTCACCACCTTTAATC | GATG | 0.333 | 0.795 | 0.958 | 0.353 |
| A17 | TET-CTCTGCTGAGACGTTAATGAT | GTAAATCGTACCCAAGAGGTT | TGG | 0.534 | 0.563 | 0.967 | 0.593 |
| A25 | FAM-GGGCATACCTCTCTTCTCTTA | GGATTGCTAGGTATGGTCTTT | AGT | 0.347 | 0.785 | 0.954 | 0.367 |
| A51 | HEX-ATTAACAATGGTCCGAAACTT | GACAAGGAGAAAGCCATTAGT | AAGC | 0.465 | 0.702 | 0.941 | 0.486 |
| A74 | TET-GATCGATTCTCGCTTAAAGAC | TCCTGCTCCACTATACTCTCA | TGC | 0.790 | 0.286 | 0.979 | 0.814 |
| A80 | FAM-GGGACATCGACAATATGTAAG | GAGCTCTGCTTTGAGACATAA | CTG | 0.552 | 0.603 | 0.937 | 0.588 |
| A83 | HEX-GAATCTTTCGGTTTAATGGTT | GGGAATGGTGGTATCATAGTA | CTTT | 0.644 | 0.532 | 0.975 | 0.671 |
| A87 | TET-GGAGAAACATCAATCAATCAA | CTGAGAGGAACCAAGATGTC | AGCA | 0.508 | 0.651 | 0.958 | 0.540 |
| A101 | FAM-ACAACATCAACTACCATCCAC | GGCTATTGGAAGAAGGATAAG | CTC | 0.811 | 0.330 | 0.925 | 0.828 |
| A103 | HEX-TCACTGCACTATCTCCAATCT | ACACGACATCGAAACATACTT | TGTA | 0.734 | 0.329 | 0.979 | 0.765 |
| A126 | TET-GCCAGGTGGTTAGGAGTATAA | TATTTGAACCACCATAACGTC | AAAG | 0.697 | 0.420 | 0.967 | 0.733 |
| A149 | FAM-AAAGAATGTGTGTCATCGAAT | TTCATCCTAGTTCCGTCAGTA | CA | 0.458 | 0.714 | 0.908 | 0.474 |
| A154 | HEX-TAATCTGAATGGTTGGTTGTT | CATGAAGGACTGTCAACTAGC | GCTA | 0.475 | 0.640 | 0.954 | 0.528 |
| A156 | TET-ATGTTTAATTTCCCTCCAAAC | TTCTCTCAACTCGCAATTCT | TGA | 0.682 | 0.475 | 0.933 | 0.712 |
| A160 | FAM-GAACATGCACGTGTGAGATA | GACTCCATACGAGACCATACA | AGTC | 0.430 | 0.726 | 0.933 | 0.452 |
| A177 | HEX-GTTCGAGGGATAGTAGTGGTT | CCAACGCATATCTTTTACCTA | AG | 0.712 | 0.358 | 0.912 | 0.747 |
| A187 | TET-GTCCAAGTTTGTCCAGTACAC | ATCGTGGAGAATATACCGAAT | CCT | 0.628 | 0.491 | 0.937 | 0.672 |
| A191 | FAM-AACACATCTCATTCATCCATC | ACCTGACATTTGACAGTTGAC | CAC | 0.858 | 0.225 | 0.950 | 0.870 |
| A192 | HEX-CCTAATACCCAACCGAATAAC | AAACAGGTGTAACTGGGTTCT | CTC | 0.541 | 0.610 | 0.933 | 0.579 |
| Population | No. of Isolates | MLST Genotype (No. of Isolates in Each Genotype) | Private MLST Genotype | Genotypic Diversity |
|---|---|---|---|---|
| HuB | 17 | 1 (3); 2 (1); 4 (1); 21 (1); 23 (4); 28 (1); 29 (1); 30 (3); 36 (2) | 4; 21; 28; 29; 30 | 0.904 |
| HeN | 8 | 1 (1); 23 (4); 24 (1); 25 (1); 50 (1) | 25; 50 | 0.786 |
| ZheJ | 5 | 1 (1); 7 (1); 16 (1); 23 (2) | 0.900 | |
| NeiM | 7 | 5 (1); 6 (2); 7 (4) | 5; 6 | 0.667 |
| ShanX | 17 | 1 (1); 2 (1); 19 (1); 20 (1); 23 (10); 24 (2); 57 (1) | 19; 20; 57 | 0.662 |
| JiL | 14 | 7 (14) | 0 | |
| QingH | 10 | 1 (1); 2 (1); 7 (2); 23 (2); 31 (1); 47 (1); 51 (1); 52 (1) | 31; 47; 51; 52 | 0.956 |
| XinJ1 | 10 | 23 (10) | 0 | |
| XinJ2 | 13 | 23 (13) | 0 | |
| GuangD | 5 | 3 (1); 7 (1); 23 (3) | 3 | 0.700 |
| GuangX | 19 | 1 (7); 2 (1); 22 (1); 23 (4); 24 (6) | 0.754 | |
| HaiN | 22 | 9 (1); 10 (1); 11 (10); 12 (1); 13 (1); 22 (1); 23 (2); 26 (1); 41 (4) | 9; 10; 11; 12; 13; 26; 41 | 0.775 |
| Dianchi_YunN | 13 | 23 (8); 45 (1); 48 (1); 49 (2); 53 (1) | 48; 49 | 0.628 |
| GeJ_YunN | 15 | 1 (3); 23 (6); 53 (2); 58 (4) | 0.286 | |
| YiM_YunN | 24 | 8 (1); 33 (1); 34 (4); 35 (1); 37 (1); 38 (1); 39 (1); 53 (1); 54 (1); 55 (1); 56 (1); 58 (9); 59 (1) | 8; 33; 35; 37; 38; 39; 54; 55; 56; 59 | 0.848 |
| HeiJ_YunN | 15 | 14 (1); 16 (1); 18 (1); 24 (1); 32 (1); 34 (1); 36 (2); 40 (1); 42 (1); 44 (1); 45 (1); 46 (1); 53 (2) | 14; 18; 32; 40; 42; 44; 46 | 0.981 |
| GuiZ | 9 | 15 (1); 16 (5); 17 (1); 27 (1); 58 (1) | 15; 17; 27 | 0.722 |
| SiC | 13 | 23 (3); 45 (10) | 0.385 | |
| Tibet | 3 | 43 (1); 58 (2) | 43 | 0.667 |
| Sample Groups | Sample Size | Phylogenetic Compatibility (p Value) | rBarD (p Value) | ||
|---|---|---|---|---|---|
| MLST | STR | MLST | STR | ||
| Total | 239 | 0 (1) | 0.01 (<0.001) | 0.6398 (<0.001) | 0.355 (<0.001) |
| Central | 25 | 0.8667 (0.447) | 0.7 (0.038) | 0.1077 (0.009) | 0.1331 (<0.001) |
| East | 5 | 1 (1) | 0.9947 (0.096) | 0.6805 (0.001) | 0.3039 (<0.001) |
| North | 24 | 0.9333 (1) | 0.8 (<0.001) | 0.2784 (<0.001) | 0.2772 (<0.001) |
| Northeast | 14 | 1 (1) | 0.9211 (0.009) | -nan * (<0.001) | 0.115 (<0.001) |
| Northwest | 33 | 0.8 (0.889) | 0.8632 (<0.001) | 0.576 (<0.001) | 0.3988 (<0.001) |
| South | 46 | 0.4667 (<0.001) | 0.2526 (<0.001) | 0.6867 (<0.001) | 0.288 (<0.001) |
| Southwest | 92 | 0.2667 (<0.001) | 0.0579 (<0.001) | 0.661 (<0.001) | 0.3874 (<0.001) |
| Clade I | 151 | 0.1333 (<0.001) | 0.021 (<0.001) | 0.5042 (<0.001) | 0.2019 (<0.001) |
| Clade II | 46 | 0.5333 (<0.001) | 0.3052 (<0.001) | 0.6379 (<0.001) | 0.4439 (<0.001) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, D.; Xu, J.; Dong, J.; Li, H.; Wang, D.; Gu, J.; Zhang, K.-Q.; Zhang, Y. Historical Differentiation and Recent Hybridization in Natural Populations of the Nematode-Trapping Fungus Arthrobotrys oligospora in China. Microorganisms 2021, 9, 1919. https://doi.org/10.3390/microorganisms9091919
Zhou D, Xu J, Dong J, Li H, Wang D, Gu J, Zhang K-Q, Zhang Y. Historical Differentiation and Recent Hybridization in Natural Populations of the Nematode-Trapping Fungus Arthrobotrys oligospora in China. Microorganisms. 2021; 9(9):1919. https://doi.org/10.3390/microorganisms9091919
Chicago/Turabian StyleZhou, Duanyong, Jianping Xu, Jianyong Dong, Haixia Li, Da Wang, Juan Gu, Ke-Qin Zhang, and Ying Zhang. 2021. "Historical Differentiation and Recent Hybridization in Natural Populations of the Nematode-Trapping Fungus Arthrobotrys oligospora in China" Microorganisms 9, no. 9: 1919. https://doi.org/10.3390/microorganisms9091919
APA StyleZhou, D., Xu, J., Dong, J., Li, H., Wang, D., Gu, J., Zhang, K.-Q., & Zhang, Y. (2021). Historical Differentiation and Recent Hybridization in Natural Populations of the Nematode-Trapping Fungus Arthrobotrys oligospora in China. Microorganisms, 9(9), 1919. https://doi.org/10.3390/microorganisms9091919







