Isolation and Characterization of Outer Membrane Vesicles of Pectobacterium brasiliense 1692
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Strains and Growth Conditions
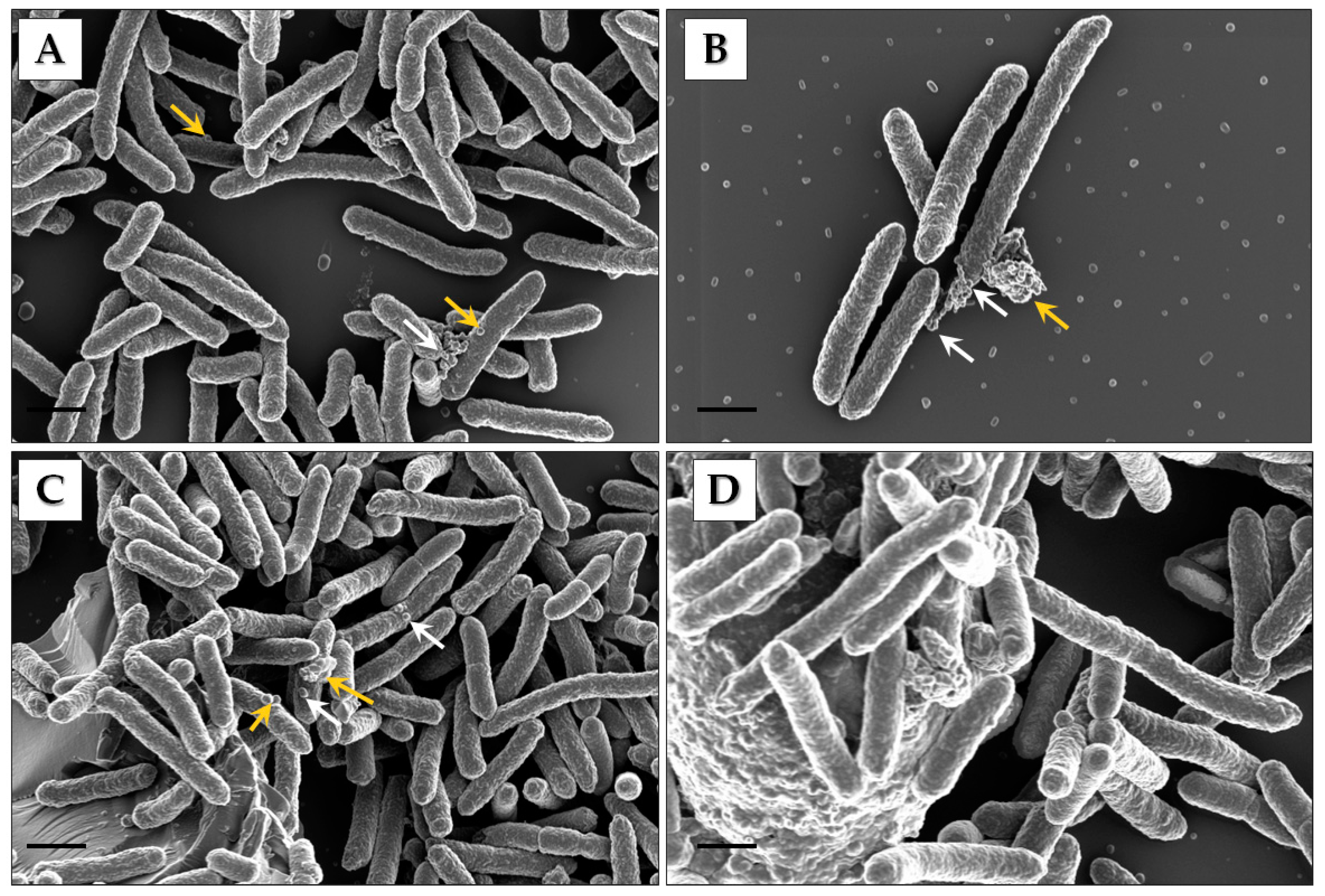
2.2. Scanning Electron Microscopy Analysis of Cells
2.3. Transmission Electron Microscopy Analysis of Cells
2.4. Isolation of OMVs
2.5. Negative Staining of OMVs
2.6. Nanoparticle Tracking Analysis and Bradford Assay of Cell-Free Supernatants
2.7. Protein Extraction and Digestion
2.8. High-Performance Liquid Chromatography (HPLC)
2.9. Electrospray Ionization Mass Spectrometry (ESI-MS/MS)
2.10. Database Search and Quantification
2.11. Sequence Analysis and Annotations
2.12. Virulence Assays of Pbr1692 OMVs
2.12.1. Protease Activity of OMVs
2.12.2. Maceration of Potato Tubers
2.12.3. Nicotiana benthamiana Elicitation of Hypersensitive Response by Pbr1692 OMVs
2.13. Antimicrobial Activity of OMVs
2.14. Statistical Analysis
3. Results
3.1. Identification of Pbr1692 OMVs
3.2. Electron Microscopy and Nanoparticle Analysis of Vesicles
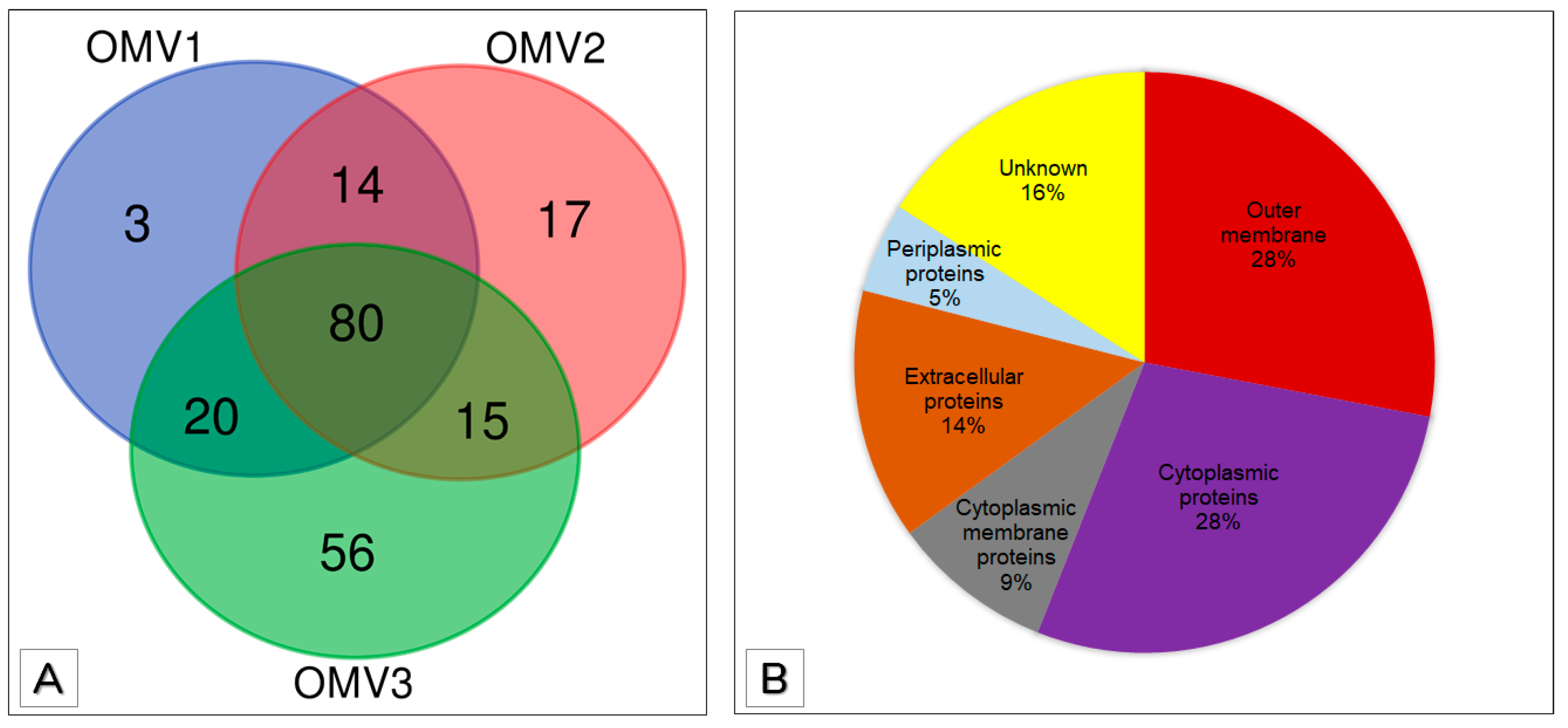
3.3. Proteomic Analysis of OMVs
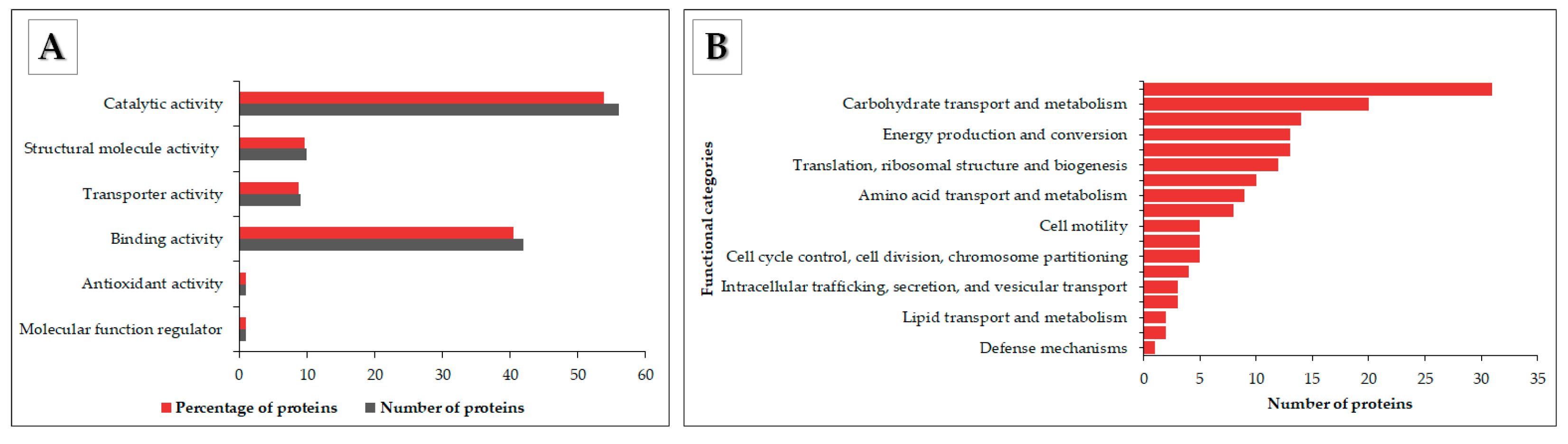
3.4. Functional Annotation of OMV Proteins
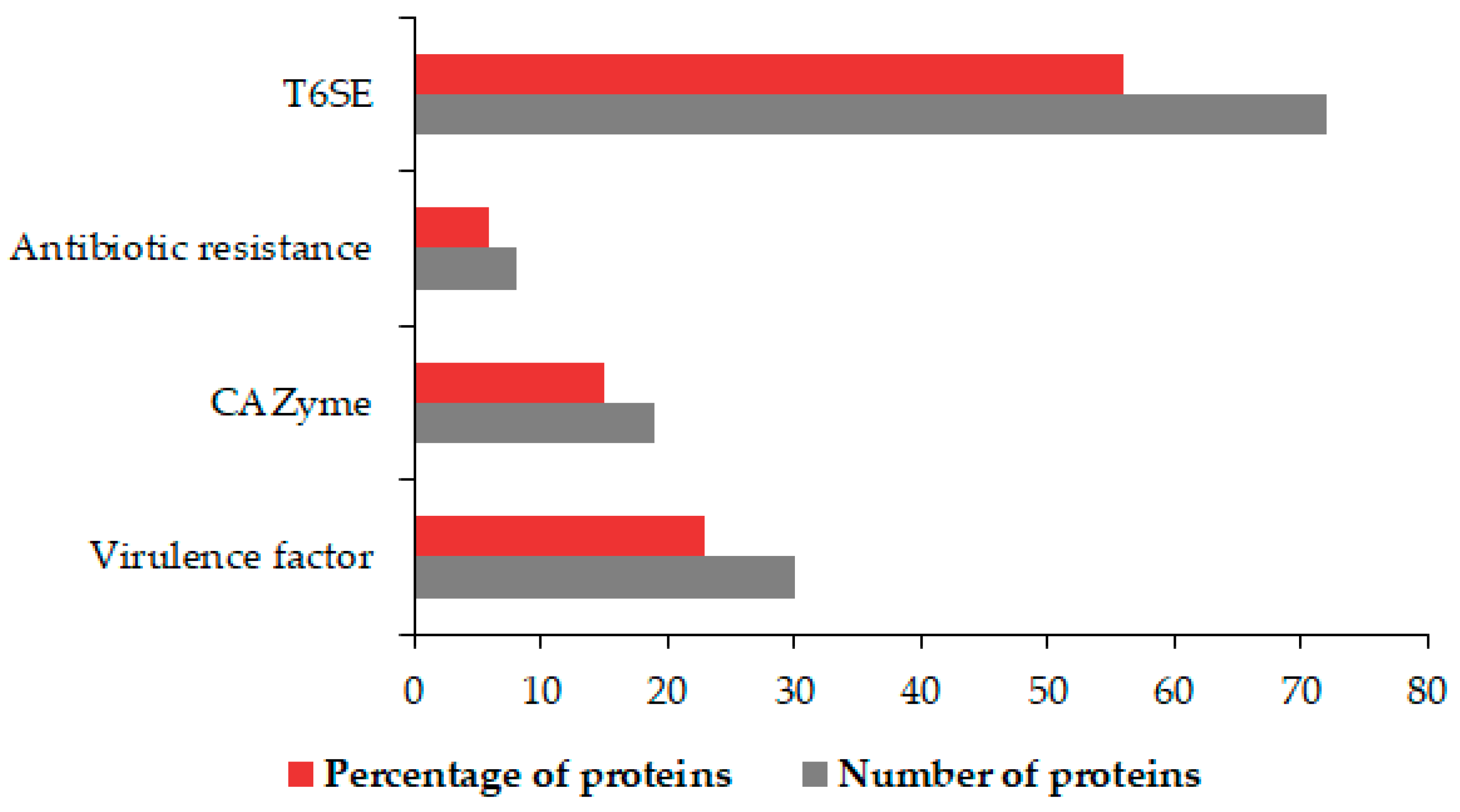
3.5. Identification of OMV Virulence Factors, Carbohydrate-Active Proteins, Antibiotic Agents, and T6SEs
3.6. Contribution of Pbr1692 OMVs to Virulence and Hypersensitive Response
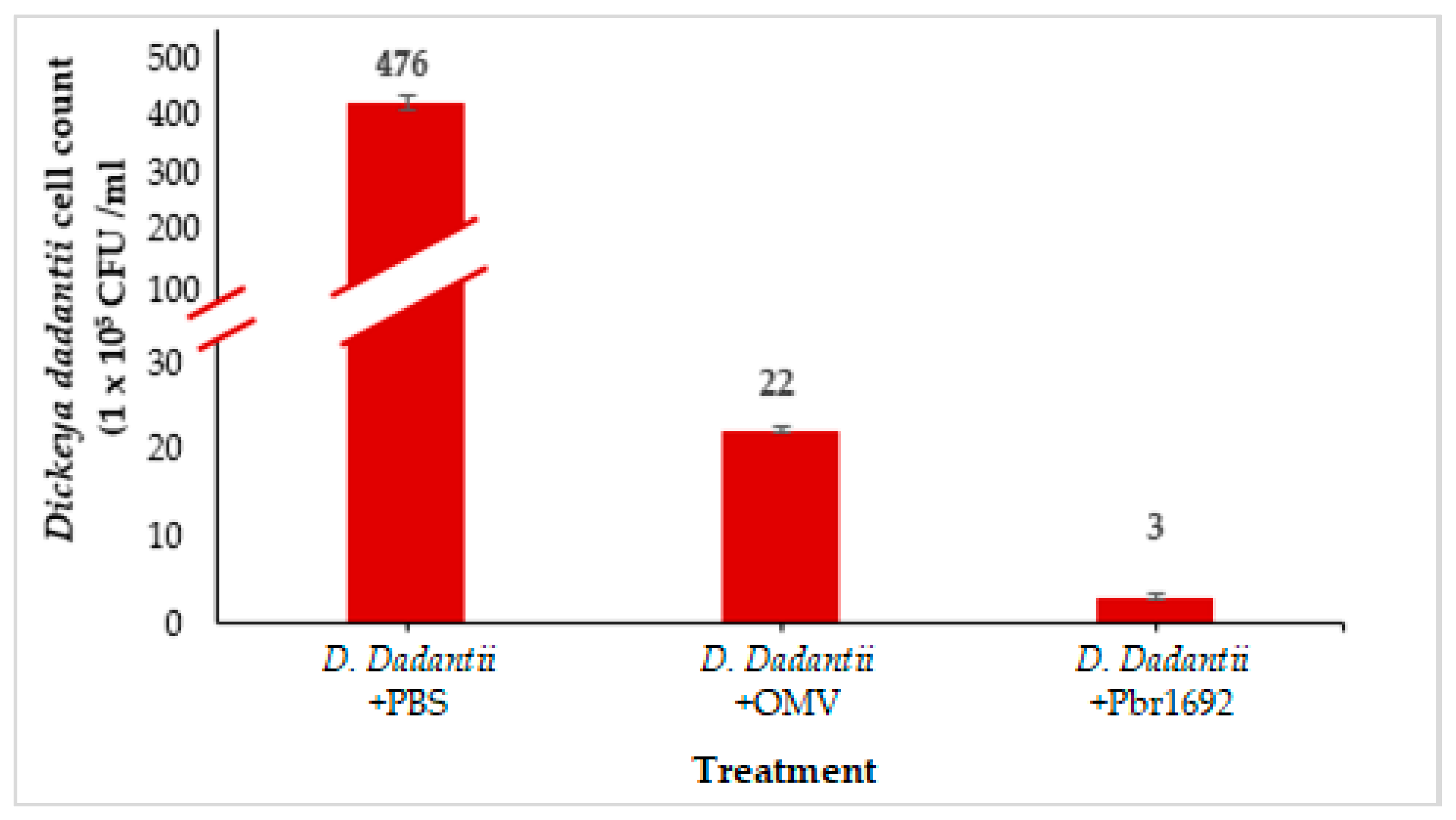
3.7. Antimicrobial Activity of OMVs against Dickeya dadantii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, J.H.; Desveaux, D.; Creason, A.L. The ABCs and 123s of bacterial secretion systems in plant pathogenesis. Annu. Rev. Phytopathol. 2014, 52, 317–345. [Google Scholar] [CrossRef]
- Charkowski, A.O. The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef]
- Zaczek-Moczydłowska, M.A.; Fleming, C.C.; Young, G.K.; Campbell, K.; O’Hanlon, R. Pectobacterium and Dickeya species detected in vegetables in Northern Ireland. Eur. J. Plant Pathol. 2019, 154, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Van der Merwe, J.J.; Coutinho, T.A.; Korsten, L.; van der Waals, J.E. Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur. J. Plant Pathol. 2010, 126, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Onkendi, E.M.; Moleleki, L.N. Characterization of Pectobacterium carotovorum subsp. carotovorum and brasiliense from diseased potatoes in Kenya. Eur. J. Plant Pathol. 2014, 139, 557–566. [Google Scholar] [CrossRef]
- Van der Wolf, J.; De Haan, E.; Kastelein, P.; Krijger, M.; De Haas, B.; Velvis, H.; Mendes, O.; Kooman-Gersmann, M.; Van Der Zouwen, P. Virulence of Pectobacterium carotovorum subsp. brasiliense on potato compared with that of other Pectobacterium and Dickeya species under climatic conditions prevailing in the Netherlands. Plant Pathol. 2017, 66, 571–583. [Google Scholar] [CrossRef]
- Guerrero-Mandujano, A.; Hernández-Cortez, C.; Ibarra, J.A.; Castro-Escarpulli, G. The outer membrane vesicles: Secretion system type zero. Traffic 2017, 18, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Davidsson, P.R.; Kariola, T.; Niemi, O.; Palva, T. Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 2013, 4, 191. [Google Scholar] [CrossRef] [Green Version]
- Fukuoka, S.; Kamishima, H.; Tamiya, E.; Karube, I. Spontaneous release of outer membrane vesicles by Erwinia carotovora. Microbios 1992, 72, 167–173. [Google Scholar]
- Yaganza, E.-S.; Rioux, D.; Simard, M.; Arul, J.; Tweddell, R.J. Ultrastructural alterations of Erwinia carotovora subsp. atroseptica caused by treatment with aluminum chloride and sodium metabisulfite. Appl. Environ. Microbiol. 2004, 70, 6800–6808. [Google Scholar] [CrossRef] [Green Version]
- Piotrowska, M.; Ciura, K.; Zalewska, M.; Dawid, M.; Correia, B.; Sawicka, P.; Lewczuk, B.; Kasprzyk, J.; Sola, L.; Piekoszewski, W. Capillary zone electrophoresis of bacterial extracellular vesicles: A proof of concept. J. Chromatogr. A 2020, 1621, 461047. [Google Scholar] [CrossRef] [PubMed]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Virulence Mech. Bact. Pathog. 2016, 4, 213–239. [Google Scholar] [CrossRef] [Green Version]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Woith, E.; Fuhrmann, G.; Melzig, M.F. Extracellular vesicles—Connecting kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [Green Version]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Roier, S.; Zingl, F.G.; Cakar, F.; Durakovic, S.; Kohl, P.; Eichmann, T.O.; Klug, L.; Gadermaier, B.; Weinzerl, K.; Prassl, R. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Azam, F.; Zhang, S. Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ. Microbiol. 2016, 18, 3850–3866. [Google Scholar] [CrossRef]
- Orench-Rivera, N.; Kuehn, M.J. Environmentally controlled bacterial vesicle-mediated export. Cell. Microbiol. 2016, 18, 1525–1536. [Google Scholar] [CrossRef] [Green Version]
- Reimer, S.L.; Beniac, D.R.; Hiebert, S.L.; Booth, T.F.; Chong, P.M.; Westmacott, G.R.; Zhanel, G.G.; Bay, D.C. Comparative Analysis of Outer Membrane Vesicle Isolation Methods with an Escherichia coli tolA Mutant Reveals a Hypervesiculating Phenotype with Outer-Inner Membrane Vesicle Content. Front. Microbiol. 2021, 12, 383. [Google Scholar] [CrossRef] [PubMed]
- Haurat, M.F.; Elhenawy, W.; Feldman, M.F. Prokaryotic membrane vesicles: New insights on biogenesis and biological roles. Biol. Chem. 2015, 396, 95–109. [Google Scholar] [CrossRef]
- Avila-Calderón, E.D.; Araiza-Villanueva, M.G.; Cancino-Diaz, J.C.; López-Villegas, E.O.; Sriranganathan, N.; Boyle, S.M.; Contreras-Rodríguez, A. Roles of bacterial membrane vesicles. Arch. Microbiol. 2015, 197, 1–10. [Google Scholar] [CrossRef]
- Ionescu, M.; Zaini, P.A.; Baccari, C.; Tran, S.; da Silva, A.M.; Lindow, S.E. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. USA 2014, 111, E3910–E3918. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Bahar, O. Bacterial outer membrane vesicles at the plant–pathogen interface. PLoS Pathog. 2017, 13, e1006306. [Google Scholar] [CrossRef] [Green Version]
- Bahar, O.; Mordukhovich, G.; Luu, D.D.; Schwessinger, B.; Daudi, A.; Jehle, A.K.; Felix, G.; Ronald, P.C. Bacterial outer membrane vesicles induce plant immune responses. Mol. Plant-Microbe Interact. 2016, 29, 374–384. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef] [Green Version]
- Koeppen, K.; Hampton, T.H.; Jarek, M.; Scharfe, M.; Gerber, S.A.; Mielcarz, D.W.; Demers, E.G.; Dolben, E.L.; Hammond, J.H.; Hogan, D.A. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 2016, 12, e1005672. [Google Scholar] [CrossRef]
- Vanaja, S.K.; Russo, A.J.; Behl, B.; Banerjee, I.; Yankova, M.; Deshmukh, S.D.; Rathinam, V.A. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 2016, 165, 1106–1119. [Google Scholar] [CrossRef] [Green Version]
- Bitto, N.J.; Chapman, R.; Pidot, S.; Costin, A.; Lo, C.; Choi, J.; D’cruze, T.; Reynolds, E.C.; Dashper, S.G.; Turnbull, L. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host–pathogen interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef] [Green Version]
- Jan, A.T. Outer membrane vesicles (OMVs) of gram-negative bacteria: A perspective update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- MacDonald, I.A.; Kuehn, M.J. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2971–2981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roszkowiak, J.; Jajor, P.; Guła, G.; Gubernator, J.; Żak, A.; Drulis-Kawa, Z.; Augustyniak, D. Interspecies outer membrane vesicles (OMVs) modulate the sensitivity of pathogenic bacteria and pathogenic yeasts to cationic peptides and serum complement. Int. J. Mol. Sci. 2019, 20, 5577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horstman, A.L.; Kuehn, M.J. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 2000, 275, 12489–12496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhenawy, W.; Debelyy, M.O.; Feldman, M.F. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 2014, 5, e00909-14. [Google Scholar] [CrossRef] [Green Version]
- Kroniger, T.; Otto, A.; Becher, D. Proteomic analysis of bacterial (outer) membrane vesicles: Progress and clinical potential. Expert Rev. Proteom. 2018, 15, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Berleman, J.E.; Allen, S.; Danielewicz, M.A.; Remis, J.P.; Gorur, A.; Cunha, J.; Hadi, M.Z.; Zusman, D.R.; Northen, T.R.; Witkowska, H.E. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front. Microbiol. 2014, 5, 474. [Google Scholar] [CrossRef]
- Marquez-Villavicencio, M.d.P.; Groves, R.L.; Charkowski, A.O. Soft rot disease severity is affected by potato physiology and Pectobacterium taxa. Plant Dis. 2011, 95, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Shyntum, D.Y.; Nkomo, N.P.; Shingange, N.L.; Gricia, A.R.; Bellieny-Rabelo, D.; Moleleki, L.N. The impact of type VI secretion system, bacteriocins and antibiotics on bacterial competition of Pectobacterium carotovorum subsp. brasiliense and the regulation of carbapenem biosynthesis by iron and the ferric-uptake regulator. Front. Microbiol. 2019, 10, 2379. [Google Scholar] [CrossRef]
- Altindis, E.; Fu, Y.; Mekalanos, J.J. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. USA 2014, 111, E1548–E1556. [Google Scholar] [CrossRef] [Green Version]
- Davies, C.; Taylor, A.J.; Elmi, A.; Winter, J.; Liaw, J.; Grabowska, A.D.; Gundogdu, O.; Wren, B.W.; Kelly, D.J.; Dorrell, N. Sodium taurocholate stimulates Campylobacter jejuni outer membrane vesicle production via down-regulation of the maintenance of lipid asymmetry pathway. Front. Cell. Infect. Microbiol. 2019, 9, 177. [Google Scholar] [CrossRef] [Green Version]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, G. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Alvarez, R.V.; Landsman, D.; Koonin, E.V. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Wang, J.; Yang, B.; Leier, A.; Marquez-Lago, T.T.; Hayashida, M.; Rocker, A.; Zhang, Y.; Akutsu, T.; Chou, K.-C.; Strugnell, R.A. Bastion6: A bioinformatics approach for accurate prediction of type VI secreted effectors. Bioinformatics 2018, 34, 2546–2555. [Google Scholar] [CrossRef]
- Deatherage, B.L.; Lara, J.C.; Bergsbaken, T.; Barrett, S.L.R.; Lara, S.; Cookson, B.T. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 2009, 72, 1395–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schooling, S.R.; Beveridge, T.J. Membrane vesicles: An overlooked component of the matrices of biofilms. J. Bacteriol. 2006, 188, 5945–5957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Fukuda, M.; Kawakami, H.; Ochiai, K.; Hanawa, T.; Kamiya, S. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seike, S.; Kobayashi, H.; Ueda, M.; Takahashi, E.; Okamoto, K.; Yamanaka, H. Outer membrane vesicles released from Aeromonas strains are involved in the biofilm formation. Front. Microbiol. 2020, 11, 613650. [Google Scholar] [CrossRef]
- Cooke, A.C.; Florez, C.; Dunshee, E.B.; Lieber, A.D.; Terry, M.L.; Light, C.J.; Schertzer, J.W. PQS-Induced Outer Membrane Vesicles Enhance Biofilm Dispersion in Pseudomonas aeruginosa. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kubheka, G.C.; Coutinho, T.A.; Moleleki, N.; Moleleki, L.N. Colonization patterns of an mCherry-tagged Pectobacterium carotovorum subsp. brasiliense strain in potato plants. Phytopathology 2013, 103, 1268–1279. [Google Scholar] [CrossRef] [Green Version]
- Moleleki, L.N.; Pretorius, R.G.; Tanui, C.K.; Mosina, G.; Theron, J. A quorum sensing-defective mutant of Pectobacterium carotovorum ssp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Mol. Plant Pathol. 2017, 18, 32–44. [Google Scholar] [CrossRef] [Green Version]
- Cooke, A.C.; Nello, A.V.; Ernst, R.K.; Schertzer, J.W. Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS ONE 2019, 14, e0212275. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Cruz, C.; Delgado, L.; López-Iglesias, C.; Mercade, E. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS ONE 2015, 10, e0116896. [Google Scholar] [CrossRef] [Green Version]
- Pï, C.; Carriï, O.; Delgado, L.; Martinez, G.; Lï, C.; Mercade, E. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: Implications for DNA content. Appl. Environ. Microbiol. 2013, 79, 1874–1881. [Google Scholar]
- Yun, S.H.; Lee, S.-Y.; Choi, C.-W.; Lee, H.; Ro, H.-J.; Jun, S.; Kwon, Y.M.; Kwon, K.K.; Kim, S.-J.; Kim, G.-H. Proteomic characterization of the outer membrane vesicle of the halophilic marine bacterium Novosphingobium pentaromativorans US6-1. J. Microbiol. 2017, 55, 56–62. [Google Scholar] [CrossRef]
- Veith, P.D.; Chen, Y.-Y.; Gorasia, D.G.; Chen, D.; Glew, M.D.; O’Brien-Simpson, N.M.; Cecil, J.D.; Holden, J.A.; Reynolds, E.C. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J. Proteome Res. 2014, 13, 2420–2432. [Google Scholar] [CrossRef]
- Lee, J.; Kim, O.Y.; Gho, Y.S. Proteomic profiling of Gram-negative bacterial outer membrane vesicles: Current perspectives. Proteom.–Clin. Appl. 2016, 10, 897–909. [Google Scholar] [CrossRef]
- Charkowski, A.; Blanco, C.; Condemine, G.; Expert, D.; Franza, T.; Hayes, C.; Hugouvieux-Cotte-Pattat, N.; Solanilla, E.L.; Low, D.; Moleleki, L. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 2012, 50, 425–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.-F.; Li, H.; Lin, X.-M.; Peng, X.-X. Outer membrane proteomics of kanamycin-resistant Escherichia coli identified MipA as a novel antibiotic resistance-related protein. FEMS Microbiol. Lett. 2015, 362, fnv074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutter, C.; Lehner, R.; Wirth, C.; Condemine, G.; Peneff, C.; Schirmer, T. Structure of the oligogalacturonate-specific KdgM porin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Grinter, R.; Leung, P.M.; Wijeyewickrema, L.C.; Littler, D.; Beckham, S.; Pike, R.N.; Walker, D.; Greening, C.; Lithgow, T. Protease-associated import systems are widespread in Gram-negative bacteria. PLoS Genet. 2019, 15, e1008435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, C.; Jagannadham, M.V. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim. Biophys. Acta-Proteins Proteom. 2013, 1834, 231–239. [Google Scholar] [CrossRef]
- Solé, M.; Scheibner, F.; Hoffmeister, A.-K.; Hartmann, N.; Hause, G.; Rother, A.; Jordan, M.; Lautier, M.; Arlat, M.; Büttner, D. Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J. Bacteriol. 2015, 197, 2879–2893. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, R.; Gouran, H.; Chakraborty, S.; Gillespie, H.W.; Almeida-Souza, H.O.; Tu, A.; Rao, B.J.; Feldstein, P.A.; Bruening, G.; Goulart, L.R. The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Sci. Rep. 2016, 6, 18598. [Google Scholar]
- McMillan, H.M.; Zebell, S.G.; Ristaino, J.B.; Dong, X.; Kuehn, M.J. Protective plant immune responses are elicited by bacterial outer membrane vesicles. Cell Rep. 2021, 34, 108645. [Google Scholar] [CrossRef] [PubMed]
- Corbett, M.; Virtue, S.; Bell, K.; Birch, P.; Burr, T.; Hyman, L.; Lilley, K.; Poock, S.; Toth, I.; Salmond, G. Identification of a new quorum-sensing-controlled virulence factor in Erwinia carotovora subsp. atroseptica secreted via the type II targeting pathway. Mol. Plant-Microbe Interact. 2005, 18, 334–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazemi-Pour, N.; Condemine, G.; Hugouvieux-Cotte-Pattat, N. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics 2004, 4, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Franza, T.; Expert, D. Iron uptake in soft rot Erwinia. In Iron Uptake and Homeostasis in Microorganisms; Cornelis, P., Andrews, S.C., Eds.; Caister Academic Press: Poole, UK, 2010; pp. 101–115. [Google Scholar]
- Aoki, S.K.; Diner, E.J.; de Roodenbeke, C.T.K.; Burgess, B.R.; Poole, S.J.; Braaten, B.A.; Jones, A.M.; Webb, J.S.; Hayes, C.S.; Cotter, P.A. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 2010, 468, 439–442. [Google Scholar] [CrossRef] [Green Version]
- Charkowski, A.O. Biology and control of Pectobacterium in potato. Am. J. Potato Res. 2015, 92, 223–229. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Jagannadham, M.V. Vesicles-mediated resistance to antibiotics in bacteria. Front. Microbiol. 2015, 6, 758. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Sample | Total Spectra | Identified Spectra | Identified Peptides | Identified Proteins |
|---|---|---|---|---|
| OMV1 | 40,254 | 1294 | 629 | 241 |
| OMV2 | 39,260 | 1474 | 685 | 262 |
| OMV3 | 39,726 | 2167 | 1002 | 364 |
| Total | 119,240 | 4935 | 2316 | 451 |
| Rank | Protein ID | Description | Molecular Weight [kDa] | Protein Length [aa] | Unique Peptide Number | Gene Ontology IDs | Final Localization | iBAQ |
|---|---|---|---|---|---|---|---|---|
| 1 | tr|A0A6I6X5L4|A0A6I6X5L4_9GAMM | Uncharacterized protein | 52.67 | 492 | 7 | No information | Unknown | 7865.322 |
| 2 | tr|A0A086EQL9|A0A086EQL9_9GAMM | MipA/OmpV family protein | 27.47 | 249 | 6 | GO:0016021 | Outer membrane | 6915.651 |
| 3 | tr|A0A0M2F0M9|A0A0M2F0M9_9GAMM | Phage tail sheath family protein | 51.19 | 475 | 11 | No information | Unknown | 6680.566 |
| 4 | tr|A0A6I6XF41|A0A6I6XF41_9GAMM | Porin | 39.82 | 369 | 12 | GO:0006811 GO:0015288 GO:0009279 GO:0046930 | Outer membrane | 4079.296 |
| 5 | tr|A0A3S0ZK54|A0A3S0ZK54_9GAMM | Lipoprotein NlpD | 35.59 | 344 | 7 | No information | Outer membrane | 2786.763 |
| 6 | tr|A0A0M2F4E7|A0A0M2F4E7_9GAMM | Avirulence protein | 68.42 | 621 | 15 | No information | Unknown | 2730.042 |
| 7 | tr|A0A6I6X6T7|A0A6I6X6T7_9GAMM | Serralysin | 51.22 | 476 | 8 | No information | Extracellular | 2408.248 |
| 8 | tr|A0A6I6X997|A0A6I6X997_9GAMM | Porin | 27.64 | 243 | 4 | No information | Outer membrane | 2390.189 |
| 9 | tr|A0A086ESP1|A0A086ESP1_9GAMM | 30S ribosomal protein S4 | 23.44 | 206 | 5 | GO:0003735 GO:0019843 GO:0006412 GO:0015935 | Cytoplasmic | 2368.547 |
| 10 | tr|A0A0M2F5U6|A0A0M2F5U6_9GAMM | Tol-Pal system protein TolB | 45.89 | 430 | 8 | GO:0017038 GO:0042597 | Periplasmic | 2326,679 |
| 11 | tr|A0A6I6WLG2|A0A6I6WLG2_9GAMM | Endolytic peptidoglycan transglycosylase RlpA | 39.08 | 372 | 9 | GO:0071555 GO:0016829 GO:0008932 GO:0000270 GO:0042834 | Extracellular | 2241.485 |
| 12 | tr|A0A0M2F1K8|A0A0M2F1K8_9GAMM | Membrane protein | 18.30 | 171 | 4 | GO:0016021 GO:0009279 | Outer membrane | 2232.414 |
| 13 | tr|A0A0M2F2F9|A0A0M2F2F9_9GAMM | Endoglucanase | 54.91 | 505 | 5 | GO:0005576 GO:0030248 GO:0008810 GO:0030245 | Extracellular | 2195.773 |
| 14 | tr|A0A3S0ZW77|A0A3S0ZW77_9GAMM | DNA protection during starvation protein | 18.47 | 167 | 2 | GO:0005737 GO:0016722 GO:0006879 GO:0030261 GO:0008199 GO:0006950 GO:0003677 | Cytoplasmic | 2130.814 |
| 15 | tr|A0A6I6X0X5|A0A6I6X0X5_9GAMM | Porin | 27.26 | 238 | 6 | No information | Outer membrane | 2073.776 |
| 16 | tr|A0A6I6WXM4|A0A6I6WXM4_9GAMM | TonB-dependent siderophore receptor | 85.60 | 782 | 17 | GO:0005506 GO:0009279 GO:0004872 GO:0015891 | Outer membrane | 2064.695 |
| 17 | tr|A0A6I6X4V7|A0A6I6X4V7_9GAMM | Long-chain fatty acid transport protein | 46.74 | 433 | 7 | No information | Outer membrane | 1905.703 |
| 18 | tr|A0A0M2F6B4|A0A0M2F6B4_9GAMM | Peptidoglycan-associated protein OS | 18.52 | 170 | 3 | GO:0016021 GO:0009279 | Outer membrane | 1732.263 |
| 19 | tr|A0A086F0V9|A0A086F0V9_9GAMM | 30S ribosomal protein S21 | 8.49 | 71 | 2 | GO:0003735 GO:0005840 GO:0016787 GO:0005829 GO:0019843 GO:0000028 GO:0044391 GO:0006412 GO:0022627 | Cytoplasmic | 1724.587 |
| 20 | tr|A0A0M2F5C1|A0A0M2F5C1_9GAMM | 60 kDa chaperonin | 57.03 | 548 | 12 | GO:0005737 GO:0042026 GO:0051082 GO:0005524 | Cytoplasmic | 1668.284 |
| 21 | tr|A0A6I6WLX4|A0A6I6WLX4_9GAMM | F5/8 type C domain-containing protein | 72.11 | 683 | 8 | No information | Extracellular | 1577.233 |
| 22 | tr|A0A0M2F2C0|A0A0M2F2C0_9GAMM | Glycine zipper 2TM domain-containing protein | 15.48 | 155 | 2 | GO:0019867 | Outer membrane | 1402.157 |
| 23 | tr|A0A6I6X4G9|A0A6I6X4G9_9GAMM | Vitamin B12 transporter BtuB | 68.71 | 625 | 11 | GO:0015235 GO:0046872 GO:0046930 GO:0006811 GO:0015288 GO:0004872 GO:0009279 | Outer membrane | 1394.331 |
| 24 | tr|A0A086EV21|A0A086EV21_9GAMM | Major outer membrane lipoprotein Lpp | 8.40 | 78 | 2 | GO:0009279 GO:0019867 | Outer membrane | 1370.143 |
| 25 | tr|A0A086EK57|A0A086EK57_9GAMM | Aspartate ammonia-lyase | 52.59 | 479 | 2 | GO:0006099 GO:0006531 GO:0008797 | Cytoplasmic | 1357.524 |
| 26 | tr|A0A086ESI7|A0A086ESI7_9GAMM | Elongation factor Tu | 43.22 | 394 | 8 | GO:0005737 GO:0003746 GO:0005622 GO:0003924 GO:0005525 | Cytoplasmic | 1357.145 |
| 27 | tr|A0A6I6X1L4|A0A6I6X1L4_9GAMM | Arabinogalactan endo-beta-1,4-galactanase | 56.21 | 507 | 4 | GO:0015926 GO:0031218 GO:0008152 | Cytoplasmic membrane | 1346.036 |
| 28 | tr|A0A086EDQ7|A0A086EDQ7_9GAMM | 50S ribosomal protein L18 OS | 12.71 | 117 | 2 | GO:0003735 GO:0019843 GO:0005840 GO:0006412 | Cytoplasmic | 1340.468 |
| 29 | tr|A0A0M2EXV1|A0A0M2EXV1_9GAMM | Baseplate protein | 20.34 | 193 | 3 | No information | Unknown | 1321.959 |
| 30 | tr|A0A6I6X2M0|A0A6I6X2M0_9GAMM | Flagellar hook-associated protein 1 | 59.66 | 564 | 5 | GO:0044780 GO:0005576 GO:0071973 GO:0009424 GO:0005198 | Extracellular | 1223.784 |
| 31 | tr|A0A0M2F0F1|A0A0M2F0F1_9GAMM | Membrane protein | 23.01 | 210 | 4 | GO:0016021 GO:0009279 | Outer membrane | 1198.896 |
| 32 | tr|A0A086EVT8|A0A086EVT8_9GAMM | Membrane protein | 19.83 | 190 | 3 | No information | Unknown | 1155.032 |
| 33 | tr|A0A6I6X7Z6|A0A6I6X7Z6_9GAMM | Phage tail protein | 71.45 | 663 | 8 | No information | Unknown | 1079.253 |
| 34 | tr|A0A433N765|A0A433N765_9GAMM | Flagellin | 30.05 | 290 | 7 | GO:0005576 GO:0071973 GO:0009420 GO:0005198 | Extracellular | 1071.265 |
| 35 | tr|A0A0M2F3M0|A0A0M2F3M0_9GAMM | Pectate lyase | 40.57 | 374 | 5 | GO:0030570 GO:0016829 GO:0000272 GO:0005576 GO:0045490 GO:0046872 | Extracellular | 979.6334 |
| 36 | tr|A0A0M2F635|A0A0M2F635_9GAMM | Membrane protein | 50.45 | 463 | 7 | GO:0005215 GO:0019867 | Outer membrane | 906.776 |
| 37 | tr|A0A6I6X7V5|A0A6I6X7V5_9GAMM | TonB-dependent receptor | 78.19 | 701 | 6 | GO:0006810 GO:0009279 GO:0004872 | Outer membrane | 887.6531 |
| 38 | tr|A0A433N6R3|A0A433N6R3_9GAMM | Flagellar hook-associated protein 2 | 50.76 | 474 | 9 | GO:0007155 GO:0005576 GO:0009424 | Extracellular | 871.272 |
| 39 | tr|A0A086EFY4|A0A086EFY4_9GAMM | 50S ribosomal protein L17 | 14.73 | 130 | 3 | GO:0003735 GO:0005840 GO:0006412 | Cytoplasmic | 870.2862 |
| 40 | tr|A0A6I6WY80|A0A6I6WY80_9GAMM | Pectate lyase | 40.45 | 375 | 5 | GO:0016829 GO:0005576 GO:0000272 | Extracellular | 836.9782 |
| 41 | tr|A0A086EHI7|A0A086EHI7_9GAMM | Membrane-bound lytic murein transglycosylase | 39.88 | 357 | 3 | GO:0008933 GO:0016998 GO:0000270 GO:0071555 GO:0009279 | Unknown | 805.2442 |
| 42 | tr|A0A0M2F7U3|A0A0M2F7U3_9GAMM | Penicillin-binding protein activator LpoA | 72.40 | 672 | 5 | GO:0031241 GO:0030234 GO:0008360 GO:0009252 | Unknown | 770.3906 |
| 43 | tr|A0A086ESF5|A0A086ESF5_9GAMM | Pectate lyase | 40.21 | 374 | 5 | GO:0030570 GO:0016829 GO:0000272 GO:0005576 GO:0045490 GO:0046872 | Extracellular | 746.1676 |
| 44 | tr|A0A0M2EW43|A0A0M2EW43_9GAMM | Phosphate-binding protein PstS | 36.84 | 346 | 5 | GO:0043190 GO:0035435 GO:0042301 | Unknown | 743.0872 |
| 45 | tr|A0A0M2F4N0|A0A0M2F4N0_9GAMM | Phospholipase A1 | 33.35 | 290 | 3 | GO:0006629 GO:0008970 GO:0016020 GO:0052739 GO:0102567 GO:0102568 GO:0004623 GO:0052740 | Outer membrane | 650.1864 |
| 46 | tr|A0A086EA76|A0A086EA76_9GAMM | Ribose-phosphate pyrophosphokinase | 34.34 | 315 | 3 | GO:0005737 GO:0016301 GO:0009156 GO:0000287 GO:0004749 GO:0009165 GO:0009116 GO:0006015 GO:0005524 | Cytoplasmic | 648.6135 |
| 47 | tr|A0A086EWE3|A0A086EWE3_9GAMM | Outer membrane protein assembly factor BamA | 89.09 | 810 | 5 | GO:0051205 GO:0016021 GO:0009279 GO:0043165 | Outer membrane | 646.6694 |
| 48 | tr|A0A433N5X5|A0A433N5X5_9GAMM | Putative lipoprotein YajI | 20.43 | 189 | 2 | No information | Cytoplasmic membrane | 628.1156 |
| 49 | tr|A0A086E9U3|A0A086E9U3_9GAMM | LPS-assembly lipoprotein LptE | 20.37 | 184 | 3 | GO:0009279 GO:0043165 | Outer membrane | 617.4362 |
| 50 | tr|A0A3S1FKD4|A0A3S1FKD4_9GAMM | Penicillin-binding protein 1B | 92.49 | 826 | 4 | GO:0009252 GO:0008955 GO:0016021 GO:0008658 GO:0071555 GO:0008360 GO:0046677 GO:0009274 GO:0008233 | Cytoplasmic membrane | 312.0136 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maphosa, S.; Moleleki, L.N. Isolation and Characterization of Outer Membrane Vesicles of Pectobacterium brasiliense 1692. Microorganisms 2021, 9, 1918. https://doi.org/10.3390/microorganisms9091918
Maphosa S, Moleleki LN. Isolation and Characterization of Outer Membrane Vesicles of Pectobacterium brasiliense 1692. Microorganisms. 2021; 9(9):1918. https://doi.org/10.3390/microorganisms9091918
Chicago/Turabian StyleMaphosa, Silindile, and Lucy Novungayo Moleleki. 2021. "Isolation and Characterization of Outer Membrane Vesicles of Pectobacterium brasiliense 1692" Microorganisms 9, no. 9: 1918. https://doi.org/10.3390/microorganisms9091918
APA StyleMaphosa, S., & Moleleki, L. N. (2021). Isolation and Characterization of Outer Membrane Vesicles of Pectobacterium brasiliense 1692. Microorganisms, 9(9), 1918. https://doi.org/10.3390/microorganisms9091918





